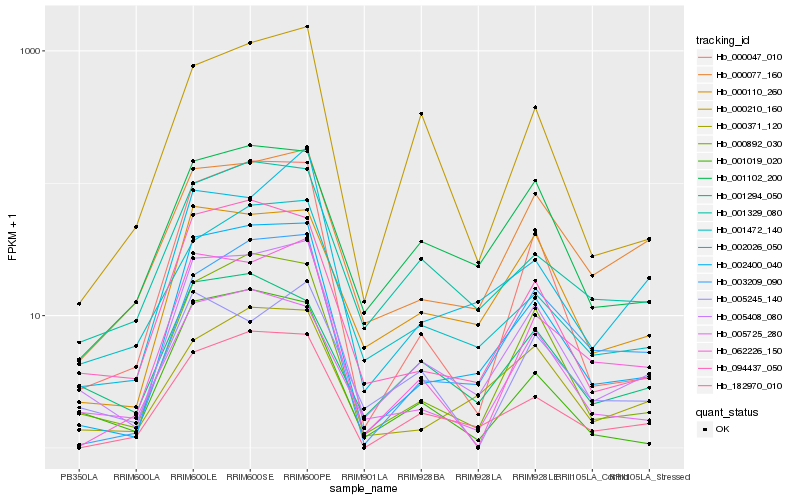
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005408_080 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 2 |
Hb_002400_040 |
0.1044158751 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 3 |
Hb_000892_030 |
0.1219118202 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 4 |
Hb_094437_050 |
0.1291340346 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000110_260 |
0.1295509583 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 6 |
Hb_005725_280 |
0.133758205 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 7 |
Hb_001329_080 |
0.1352007858 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 8 |
Hb_001472_140 |
0.1352179141 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 9 |
Hb_001102_200 |
0.1377144777 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 10 |
Hb_182970_010 |
0.1393663203 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 11 |
Hb_000077_160 |
0.139571525 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 12 |
Hb_000047_010 |
0.1417691256 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 13 |
Hb_062226_150 |
0.1427066518 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000210_160 |
0.1433019041 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_002026_050 |
0.1494551286 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 16 |
Hb_000371_120 |
0.1509194794 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_001019_020 |
0.1522969885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001294_050 |
0.153861033 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_003209_090 |
0.1540204221 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 20 |
Hb_005245_140 |
0.1548333785 |
- |
- |
PREDICTED: ribonuclease 2 [Jatropha curcas] |