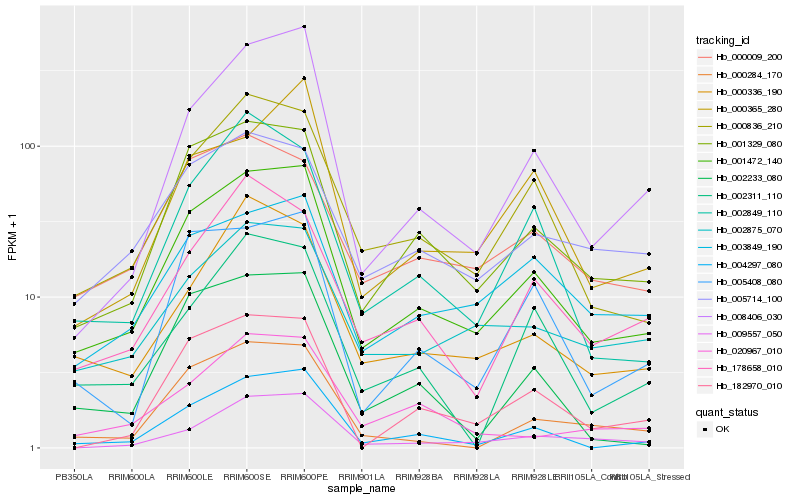
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001472_140 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 2 |
Hb_001329_080 |
0.0856648975 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 3 |
Hb_000836_210 |
0.1060329637 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 4 |
Hb_004297_080 |
0.1181609983 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_008406_030 |
0.1209922751 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 6 |
Hb_002875_070 |
0.1288390893 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 7 |
Hb_005408_080 |
0.1352179141 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 8 |
Hb_000336_190 |
0.1395502349 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 9 |
Hb_182970_010 |
0.142267478 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 10 |
Hb_000009_200 |
0.1427612865 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 11 |
Hb_005714_100 |
0.1439007849 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 12 |
Hb_178658_010 |
0.1458999616 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 13 |
Hb_009557_050 |
0.1467421231 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 14 |
Hb_003849_190 |
0.1467755401 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 15 |
Hb_002233_080 |
0.1479419137 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 16 |
Hb_000284_170 |
0.1488287226 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002849_110 |
0.1489864509 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 18 |
Hb_020967_010 |
0.1503680423 |
- |
- |
- |
| 19 |
Hb_002311_110 |
0.1538364617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000365_280 |
0.1553826984 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |