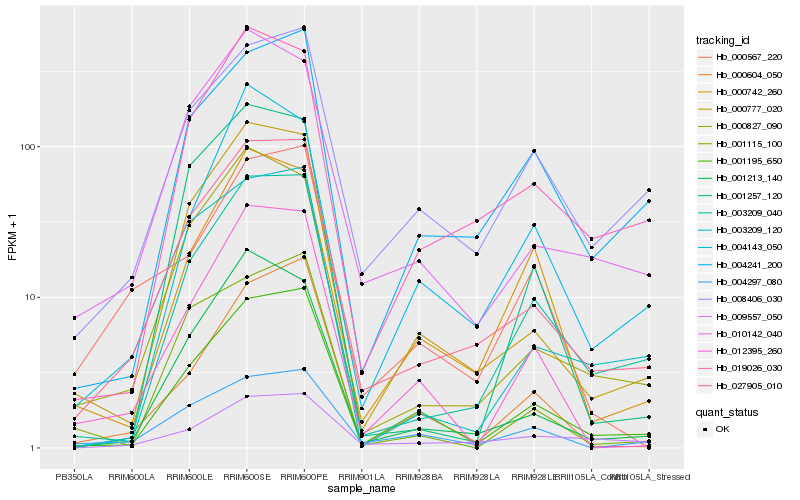
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027905_010 |
0.0 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 2 |
Hb_000777_020 |
0.0989833838 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 3 |
Hb_003209_040 |
0.112068005 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 4 |
Hb_001195_650 |
0.1126742956 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000827_090 |
0.1173509392 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 6 |
Hb_010142_040 |
0.1217348904 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 7 |
Hb_019026_030 |
0.1253123812 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 8 |
Hb_001213_140 |
0.1265471118 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 9 |
Hb_008406_030 |
0.1309330761 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 10 |
Hb_003209_120 |
0.1353652496 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 11 |
Hb_004241_200 |
0.142511914 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 12 |
Hb_012395_260 |
0.1440744081 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004297_080 |
0.1546318055 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 14 |
Hb_009557_050 |
0.1597476408 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 15 |
Hb_000604_050 |
0.16056745 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 16 |
Hb_000567_220 |
0.1610238077 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 17 |
Hb_001257_120 |
0.1621421031 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 18 |
Hb_004143_050 |
0.162845803 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 19 |
Hb_000742_260 |
0.1654230354 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 20 |
Hb_001115_100 |
0.1660825556 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |