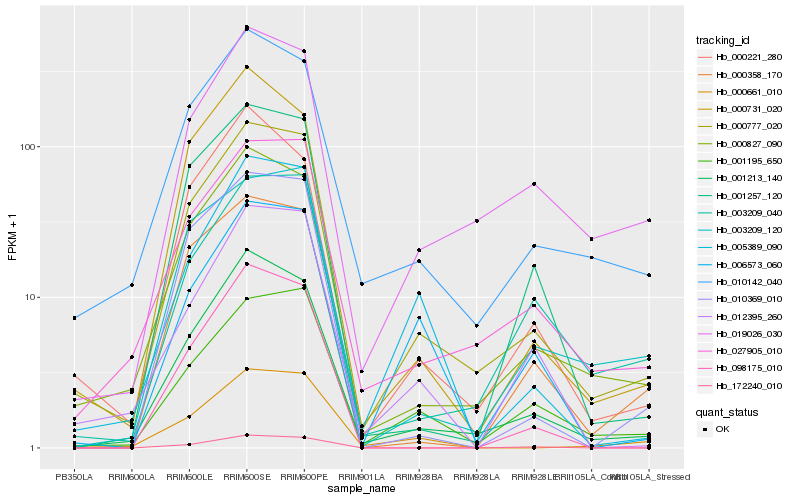
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000777_020 |
0.0 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 2 |
Hb_001213_140 |
0.0760589689 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 3 |
Hb_000827_090 |
0.0903319087 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 4 |
Hb_027905_010 |
0.0989833838 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 5 |
Hb_010142_040 |
0.1100288605 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 6 |
Hb_001195_650 |
0.1206389445 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 7 |
Hb_005389_090 |
0.1288255771 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 8 |
Hb_012395_260 |
0.1311073023 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001257_120 |
0.1324502163 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 10 |
Hb_000221_280 |
0.1325885675 |
- |
- |
PREDICTED: protein EXORDIUM-like [Populus euphratica] |
| 11 |
Hb_019026_030 |
0.1343476242 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 12 |
Hb_000661_010 |
0.1349067762 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 13 |
Hb_010369_010 |
0.1372768996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000358_170 |
0.1379776476 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 15 |
Hb_003209_040 |
0.1380291972 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 16 |
Hb_003209_120 |
0.1389061591 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 17 |
Hb_000731_020 |
0.1408830835 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 18 |
Hb_098175_010 |
0.1433009367 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 19 |
Hb_006573_060 |
0.1477617461 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 20 |
Hb_172240_010 |
0.1485116747 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |