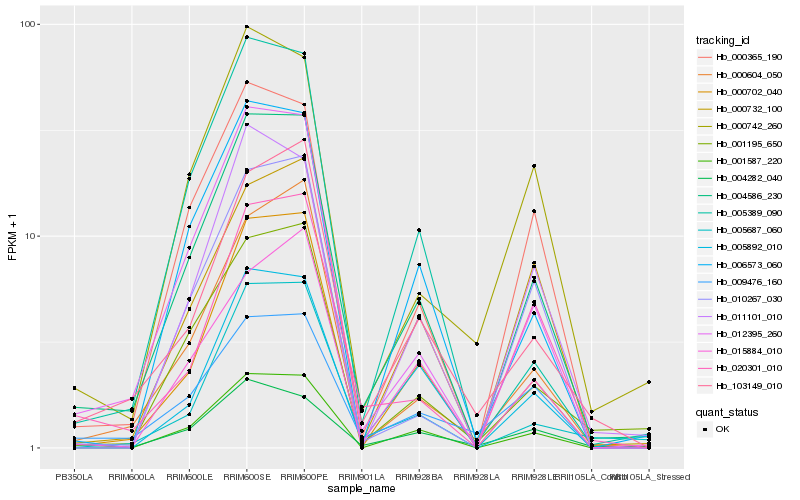
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_230 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012395_260 |
0.0834905678 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000702_040 |
0.101619026 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 4 |
Hb_000365_190 |
0.1079921186 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 5 |
Hb_001587_220 |
0.1089858113 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_006573_060 |
0.1154420108 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 7 |
Hb_001195_650 |
0.1161746714 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000604_050 |
0.1264930821 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 9 |
Hb_103149_010 |
0.1282804896 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 10 |
Hb_011101_010 |
0.1304935935 |
- |
- |
hypothetical protein JCGZ_25132 [Jatropha curcas] |
| 11 |
Hb_000732_100 |
0.1317482461 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 12 |
Hb_000742_260 |
0.1332357722 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 13 |
Hb_005687_060 |
0.1333409516 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_020301_010 |
0.1350610816 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_015884_010 |
0.1372888461 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005892_010 |
0.1390254571 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 17 |
Hb_010267_030 |
0.1420270973 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_004282_040 |
0.1423373447 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_009476_160 |
0.142558595 |
- |
- |
RING-H2 finger protein ATL2N, putative [Ricinus communis] |
| 20 |
Hb_005389_090 |
0.142892954 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |