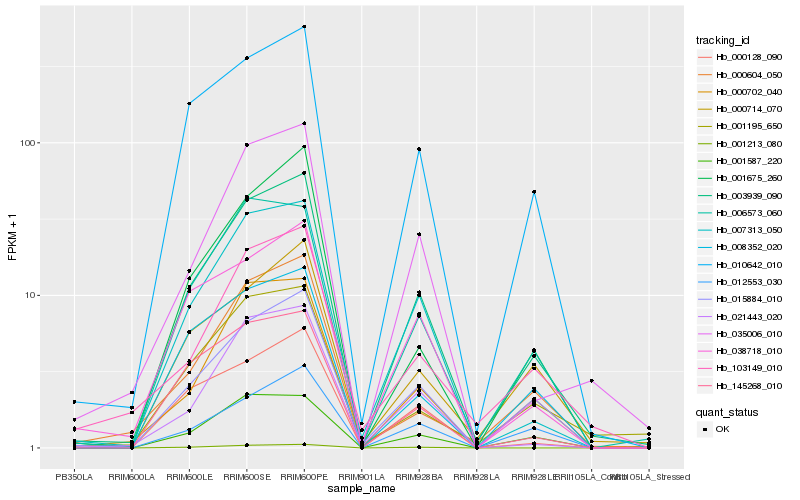
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003939_090 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_015884_010 |
0.0899552494 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007313_050 |
0.0965215951 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 4 |
Hb_010642_010 |
0.0969716523 |
- |
- |
PREDICTED: aquaporin TIP2-1 [Jatropha curcas] |
| 5 |
Hb_000702_040 |
0.11280494 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 6 |
Hb_035006_010 |
0.1130204117 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 7 |
Hb_021443_020 |
0.1180153746 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 8 |
Hb_006573_060 |
0.1183394294 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 9 |
Hb_000128_090 |
0.1192404722 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_001213_080 |
0.124051226 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 11 |
Hb_001587_220 |
0.1256601118 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000714_070 |
0.1283748867 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 13 |
Hb_145268_010 |
0.1297211841 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Jatropha curcas] |
| 14 |
Hb_038718_010 |
0.1303865087 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 15 |
Hb_001675_260 |
0.1309157494 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_008352_020 |
0.1316901573 |
- |
- |
hypothetical protein [Cecembia lonarensis] |
| 17 |
Hb_000604_050 |
0.1339194463 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 18 |
Hb_012553_030 |
0.1339347696 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 19 |
Hb_001195_650 |
0.1346193111 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 20 |
Hb_103149_010 |
0.1346973462 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |