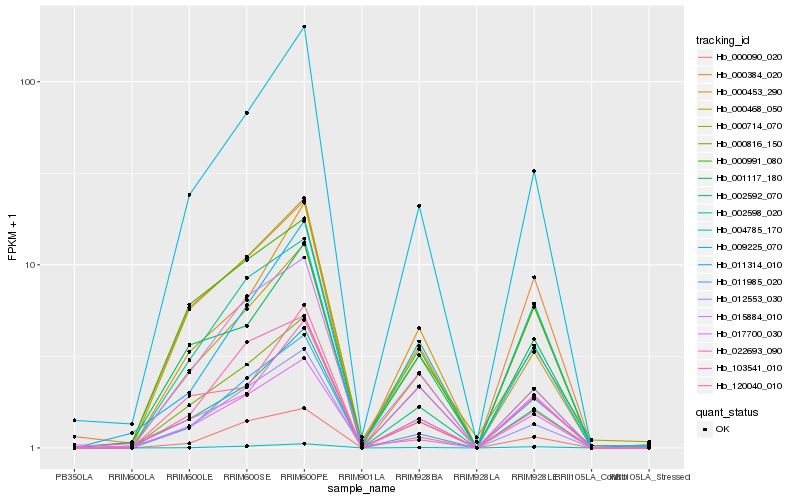
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000468_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_009225_070 |
0.0562978517 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 3 |
Hb_012553_030 |
0.0656643553 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 4 |
Hb_000816_150 |
0.0692329711 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 5 |
Hb_002592_070 |
0.0732585849 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 6 |
Hb_000453_290 |
0.0858717556 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 7 |
Hb_004785_170 |
0.1041305935 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 8 |
Hb_000090_020 |
0.1072257626 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001117_180 |
0.1083634775 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 10 |
Hb_022693_090 |
0.1117257617 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_002598_020 |
0.112000308 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 12 |
Hb_011314_010 |
0.1127331711 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000384_020 |
0.1137036077 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 14 |
Hb_000991_080 |
0.1147390743 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_015884_010 |
0.1151322984 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000714_070 |
0.1168292117 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 17 |
Hb_120040_010 |
0.1180887419 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_103541_010 |
0.1182070063 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X2 [Jatropha curcas] |
| 19 |
Hb_011985_020 |
0.1189864369 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_017700_030 |
0.119265208 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |