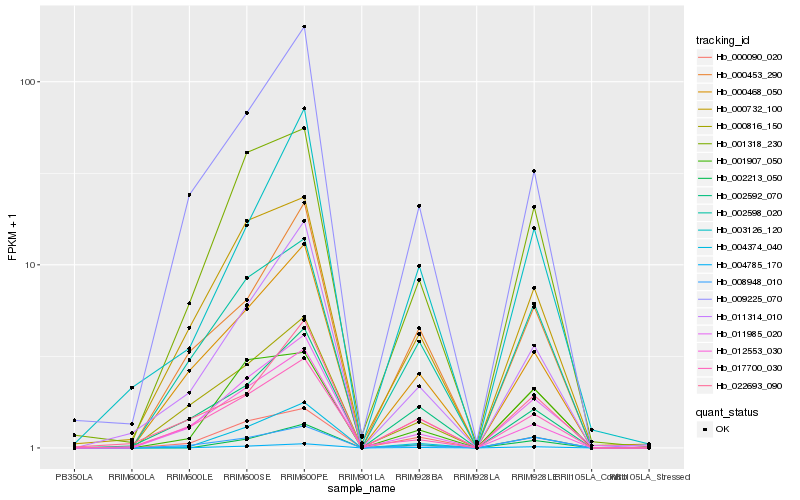
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_170 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 2 |
Hb_004374_040 |
0.0781559462 |
- |
- |
PREDICTED: beta-carotene 3-hydroxylase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_008948_010 |
0.0880555437 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011985_020 |
0.0935103184 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000468_050 |
0.1041305935 |
- |
- |
- |
| 6 |
Hb_011314_010 |
0.1099434098 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001318_230 |
0.1107046375 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 8 |
Hb_000816_150 |
0.1124491974 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 9 |
Hb_000090_020 |
0.1164965263 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 10 |
Hb_009225_070 |
0.118155496 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 11 |
Hb_002598_020 |
0.1187458371 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 12 |
Hb_000453_290 |
0.1239113504 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 13 |
Hb_002213_050 |
0.1286789375 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X3 [Jatropha curcas] |
| 14 |
Hb_017700_030 |
0.1322259214 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 15 |
Hb_022693_090 |
0.1338616633 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002592_070 |
0.1370456832 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 17 |
Hb_000732_100 |
0.1374157294 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 18 |
Hb_001907_050 |
0.137858614 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 19 |
Hb_012553_030 |
0.1378648054 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 20 |
Hb_003126_120 |
0.1400768339 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |