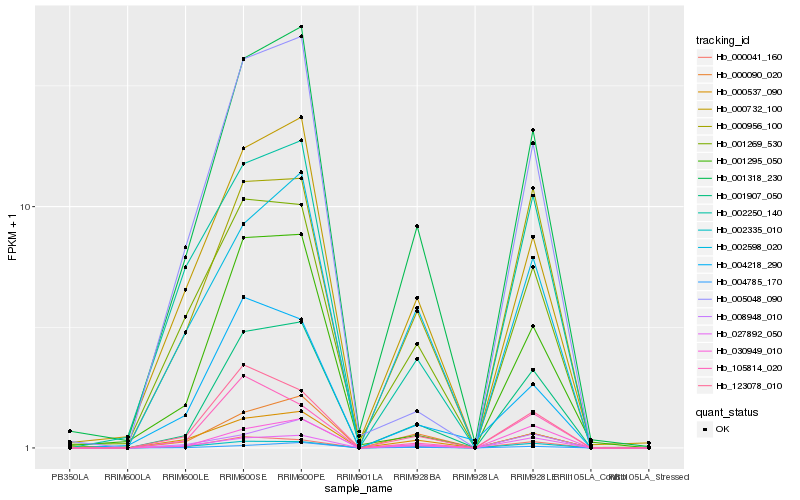
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001907_050 |
0.0 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 2 |
Hb_001318_230 |
0.0852927685 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 3 |
Hb_000041_160 |
0.1089931375 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_030949_010 |
0.1222735993 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 5 |
Hb_002335_010 |
0.1255649623 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 6 |
Hb_123078_010 |
0.132651553 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 7 |
Hb_027892_050 |
0.1331003491 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 8 |
Hb_000537_090 |
0.1366047058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000956_100 |
0.1368293602 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 10 |
Hb_000732_100 |
0.1377902356 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 11 |
Hb_004785_170 |
0.137858614 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 12 |
Hb_001295_050 |
0.1425727048 |
- |
- |
PREDICTED: uncharacterized protein LOC101503108 isoform X1 [Cicer arietinum] |
| 13 |
Hb_008948_010 |
0.1444297243 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005048_090 |
0.1470566128 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 15 |
Hb_002598_020 |
0.1492234664 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 16 |
Hb_001269_530 |
0.1524171476 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_002250_140 |
0.1525143153 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 18 |
Hb_000090_020 |
0.1553330024 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_004218_290 |
0.1570751213 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 6 isoform X2 [Jatropha curcas] |
| 20 |
Hb_105814_020 |
0.1578364196 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |