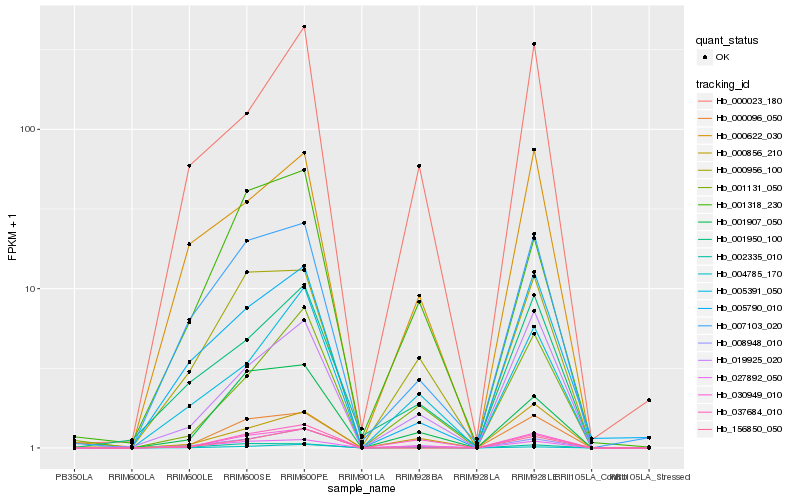
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030949_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 2 |
Hb_027892_050 |
0.0992621994 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 3 |
Hb_008948_010 |
0.1164965094 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000956_100 |
0.1211295397 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_001907_050 |
0.1222735993 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 6 |
Hb_019925_020 |
0.1323142081 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 7 |
Hb_007103_020 |
0.1335707287 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 8 |
Hb_000096_050 |
0.1468754221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000856_210 |
0.1477091994 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 10 |
Hb_001131_050 |
0.1482459558 |
- |
- |
hypothetical protein EUGRSUZ_C02968 [Eucalyptus grandis] |
| 11 |
Hb_037684_010 |
0.1514085553 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 12 |
Hb_002335_010 |
0.153441199 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 13 |
Hb_001950_100 |
0.1557081468 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 14 |
Hb_005391_050 |
0.1559721205 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 15 |
Hb_005790_010 |
0.156952357 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |
| 16 |
Hb_000023_180 |
0.1590121624 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 17 |
Hb_001318_230 |
0.1602937905 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 18 |
Hb_156850_050 |
0.160695241 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |
| 19 |
Hb_000622_030 |
0.1620745386 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 20 |
Hb_004785_170 |
0.1633470311 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |