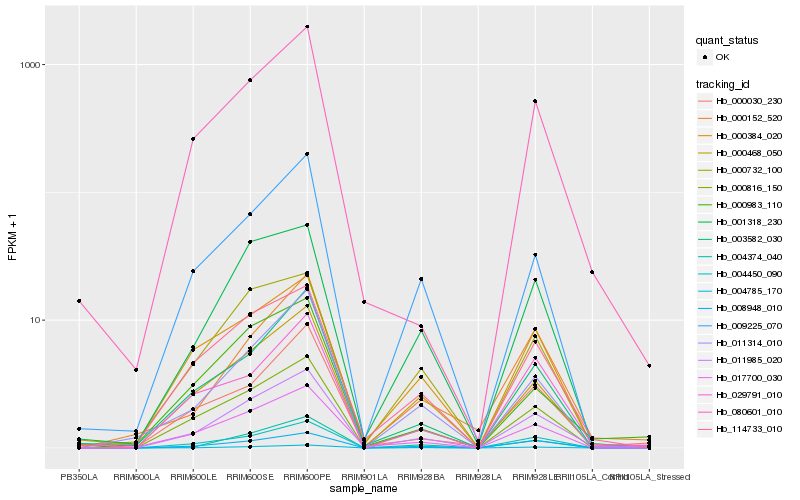
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011985_020 |
0.0 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004785_170 |
0.0935103184 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_000816_150 |
0.1049995606 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 4 |
Hb_008948_010 |
0.1100822211 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 5 |
Hb_017700_030 |
0.112866986 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 6 |
Hb_004450_090 |
0.1158707582 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 7 |
Hb_114733_010 |
0.116995499 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 8 |
Hb_000468_050 |
0.1189864369 |
- |
- |
- |
| 9 |
Hb_001318_230 |
0.1251747288 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 10 |
Hb_009225_070 |
0.1269545152 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 11 |
Hb_004374_040 |
0.1278779431 |
- |
- |
PREDICTED: beta-carotene 3-hydroxylase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_003582_030 |
0.1288738351 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 13 |
Hb_000732_100 |
0.1303087423 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 14 |
Hb_011314_010 |
0.1305119873 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000983_110 |
0.1349314168 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 16 |
Hb_000384_020 |
0.1360483926 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 17 |
Hb_029791_010 |
0.1364082729 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 18 |
Hb_080601_010 |
0.1368702147 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 19 |
Hb_000030_230 |
0.1402308833 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 20 |
Hb_000152_520 |
0.1417994249 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |