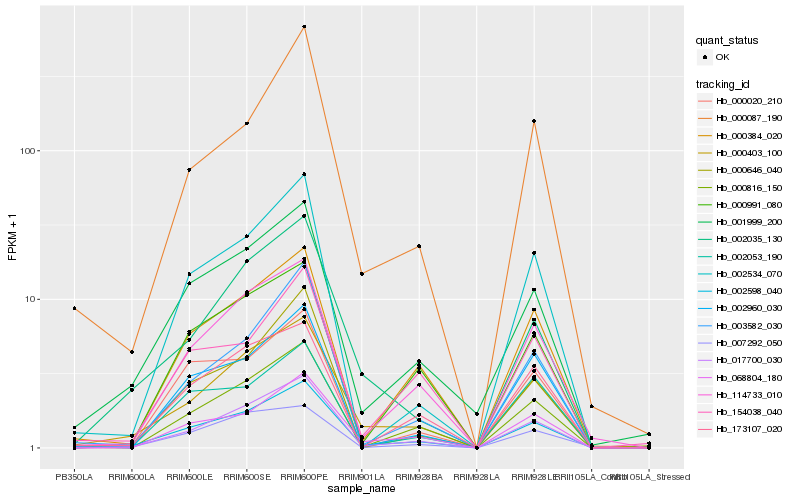
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002598_040 |
0.0 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 2 |
Hb_017700_030 |
0.0587293767 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 3 |
Hb_000403_100 |
0.0871006612 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 4 |
Hb_068804_180 |
0.1012235734 |
- |
- |
PREDICTED: TRAF-type zinc finger domain-containing protein 1 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_001999_200 |
0.102134694 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_114733_010 |
0.102781357 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 7 |
Hb_000816_150 |
0.1068474285 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 8 |
Hb_000384_020 |
0.112451359 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 9 |
Hb_002534_070 |
0.1205294269 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 10 |
Hb_000991_080 |
0.1206294628 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_002960_030 |
0.1268168776 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 12 |
Hb_173107_020 |
0.1283454802 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 13 |
Hb_000020_210 |
0.1295300422 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 14 |
Hb_003582_030 |
0.1299681885 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 15 |
Hb_002035_130 |
0.1307653126 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 16 |
Hb_000087_190 |
0.1353311405 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 17 |
Hb_000646_040 |
0.1373053095 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 18 |
Hb_154038_040 |
0.1395761102 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 19 |
Hb_002053_190 |
0.1408309766 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007292_050 |
0.1411147537 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |