| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
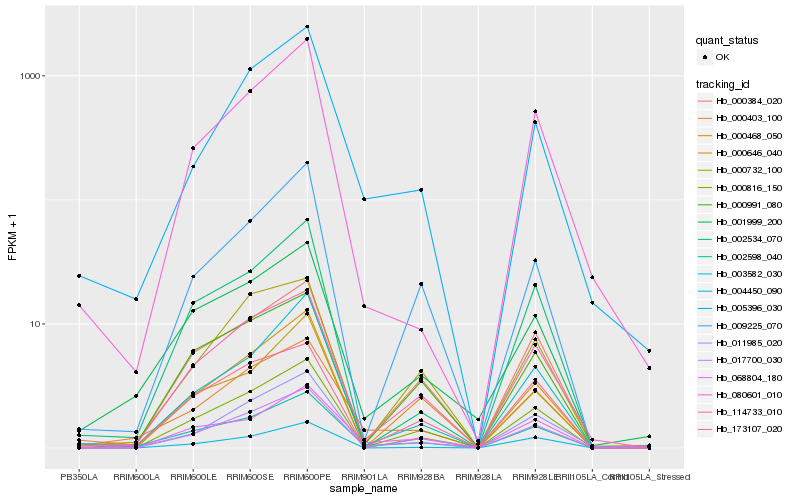
Hb_017700_030 |
0.0 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 2 |
Hb_002598_040 |
0.0587293767 |
- |
- |
hypothetical protein OsI_22030 [Oryza sativa Indica Group] |
| 3 |
Hb_000816_150 |
0.0813006328 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 4 |
Hb_114733_010 |
0.0868898884 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_000403_100 |
0.0974604594 |
- |
- |
PREDICTED: ras-related protein Rab7 [Jatropha curcas] |
| 6 |
Hb_000384_020 |
0.1097145085 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 7 |
Hb_003582_030 |
0.1097394495 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 8 |
Hb_011985_020 |
0.112866986 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002534_070 |
0.1152749511 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 10 |
Hb_004450_090 |
0.1159463765 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 11 |
Hb_009225_070 |
0.1175399046 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_005396_030 |
0.1179531272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_068804_180 |
0.1186981565 |
- |
- |
PREDICTED: TRAF-type zinc finger domain-containing protein 1 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000468_050 |
0.119265208 |
- |
- |
- |
| 15 |
Hb_000991_080 |
0.1236896534 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_173107_020 |
0.1237248334 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 17 |
Hb_000732_100 |
0.1241085342 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 18 |
Hb_000646_040 |
0.1276148781 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 19 |
Hb_080601_010 |
0.1295790583 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 20 |
Hb_001999_200 |
0.1297533894 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |