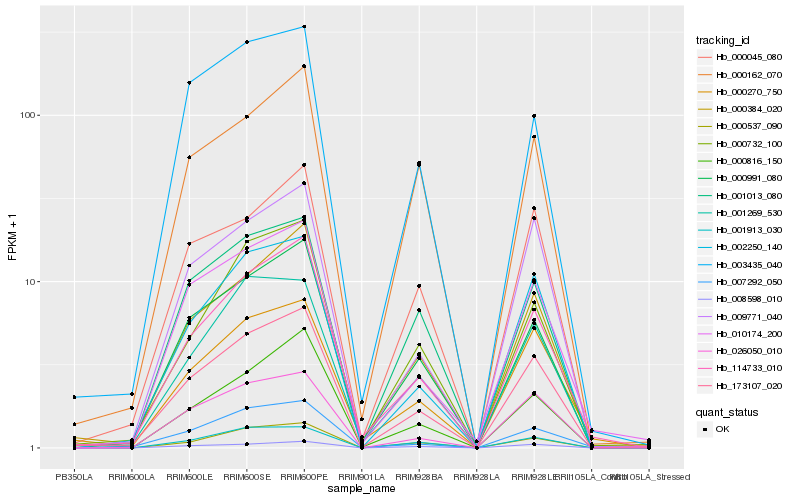
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173107_020 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 2 |
Hb_000384_020 |
0.0669777469 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 3 |
Hb_114733_010 |
0.074727972 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 4 |
Hb_002250_140 |
0.0764962253 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 5 |
Hb_000991_080 |
0.0792135069 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_009771_040 |
0.085242892 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 7 |
Hb_000045_080 |
0.0956889043 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000816_150 |
0.0979358193 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 9 |
Hb_000270_750 |
0.0981084746 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 10 |
Hb_000537_090 |
0.0982382972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008598_010 |
0.098980798 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 12 |
Hb_026050_010 |
0.1009323032 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_001269_530 |
0.1013578505 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_010174_200 |
0.1015950123 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000732_100 |
0.1019947153 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 16 |
Hb_001013_080 |
0.1045028541 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 17 |
Hb_007292_050 |
0.1056375558 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 18 |
Hb_001913_030 |
0.108234056 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 19 |
Hb_000162_070 |
0.1100833173 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 20 |
Hb_003435_040 |
0.1133197559 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |