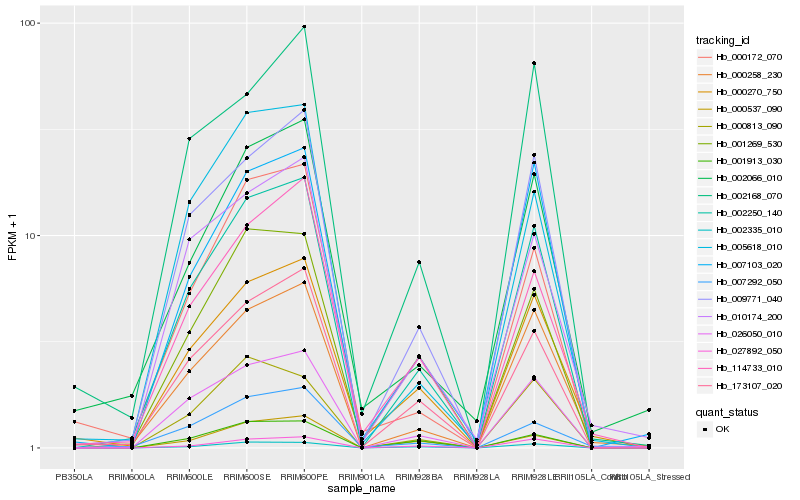
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002250_140 |
0.0 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 2 |
Hb_009771_040 |
0.073174485 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 3 |
Hb_026050_010 |
0.073310343 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 4 |
Hb_173107_020 |
0.0764962253 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 5 |
Hb_000258_230 |
0.0820659401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007103_020 |
0.1029740934 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 7 |
Hb_002335_010 |
0.1048594729 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 8 |
Hb_001269_530 |
0.1077248314 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_027892_050 |
0.109854085 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 10 |
Hb_000270_750 |
0.1100561399 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 11 |
Hb_005618_010 |
0.1133319827 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 12 |
Hb_010174_200 |
0.1153652147 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 13 |
Hb_114733_010 |
0.1159924206 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 14 |
Hb_000813_090 |
0.1173850935 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 15 |
Hb_000172_070 |
0.1185638878 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_002168_070 |
0.1207565468 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_007292_050 |
0.121119321 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 18 |
Hb_000537_090 |
0.1216528877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001913_030 |
0.1222861469 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_002066_010 |
0.1245654329 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |