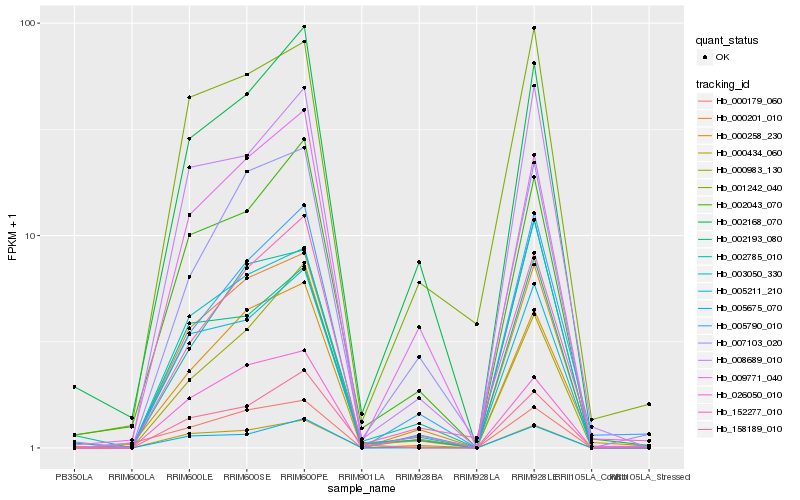
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000201_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_005211_210 |
0.0800367971 |
- |
- |
lysine and histidine specific transporter family protein [Populus trichocarpa] |
| 3 |
Hb_008689_010 |
0.0974098971 |
- |
- |
hypothetical protein POPTR_0006s25330g [Populus trichocarpa] |
| 4 |
Hb_000258_230 |
0.0991131386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002193_080 |
0.1006657134 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_002043_070 |
0.1036505503 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 7 |
Hb_003050_330 |
0.1056515321 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 8 |
Hb_005675_070 |
0.1069346986 |
- |
- |
hypothetical protein EUGRSUZ_K01585 [Eucalyptus grandis] |
| 9 |
Hb_000434_060 |
0.1085175885 |
desease resistance |
Gene Name: hmm |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 10 |
Hb_009771_040 |
0.1089334296 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 11 |
Hb_158189_010 |
0.1109286965 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 12 |
Hb_007103_020 |
0.1122339342 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 13 |
Hb_152277_010 |
0.1156627677 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_002168_070 |
0.1161201578 |
- |
- |
Jasmonate O-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000179_060 |
0.1199481307 |
- |
- |
hypothetical protein POPTR_0010s23130g, partial [Populus trichocarpa] |
| 16 |
Hb_001242_040 |
0.1237668427 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 17 |
Hb_026050_010 |
0.1242730683 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_005790_010 |
0.1268497939 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |
| 19 |
Hb_002785_010 |
0.1297528933 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 20 |
Hb_000983_130 |
0.1318334866 |
- |
- |
transcription factor, putative [Ricinus communis] |