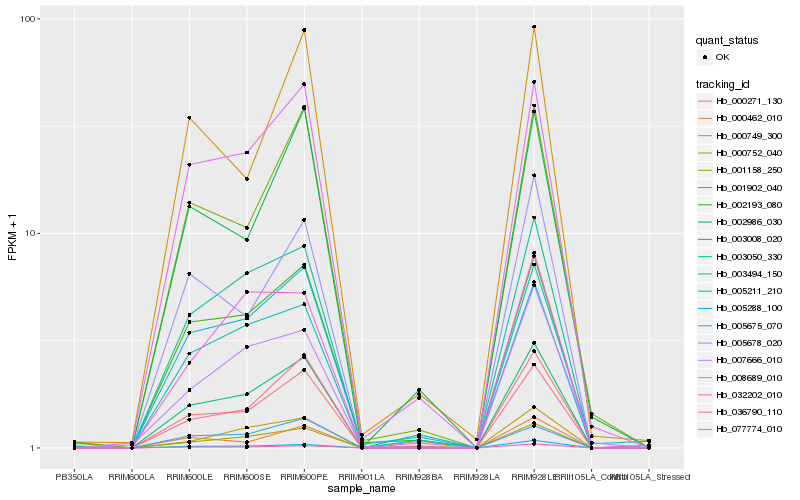
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003008_020 |
0.0 |
- |
- |
auxin-responsive family protein [Populus trichocarpa] |
| 2 |
Hb_003050_330 |
0.0792005946 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 3 |
Hb_008689_010 |
0.0828314782 |
- |
- |
hypothetical protein POPTR_0006s25330g [Populus trichocarpa] |
| 4 |
Hb_036790_110 |
0.0886465135 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 5 |
Hb_077774_010 |
0.0898913659 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_002193_080 |
0.0918375926 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_000752_040 |
0.0970728384 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_001902_040 |
0.1048648651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005288_100 |
0.1074854657 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 10 |
Hb_005678_020 |
0.1100705743 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000462_010 |
0.1102693699 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 12 |
Hb_005675_070 |
0.11222881 |
- |
- |
hypothetical protein EUGRSUZ_K01585 [Eucalyptus grandis] |
| 13 |
Hb_003494_150 |
0.1122445518 |
- |
- |
Silicon transporter, putative [Ricinus communis] |
| 14 |
Hb_000271_130 |
0.1174947474 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 15 |
Hb_007666_010 |
0.1210505117 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 16 |
Hb_032202_010 |
0.1228886113 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 17 |
Hb_001158_250 |
0.124209777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005211_210 |
0.1242376048 |
- |
- |
lysine and histidine specific transporter family protein [Populus trichocarpa] |
| 19 |
Hb_002986_030 |
0.1280966895 |
- |
- |
hypothetical protein JCGZ_12380 [Jatropha curcas] |
| 20 |
Hb_000749_300 |
0.1284368645 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |