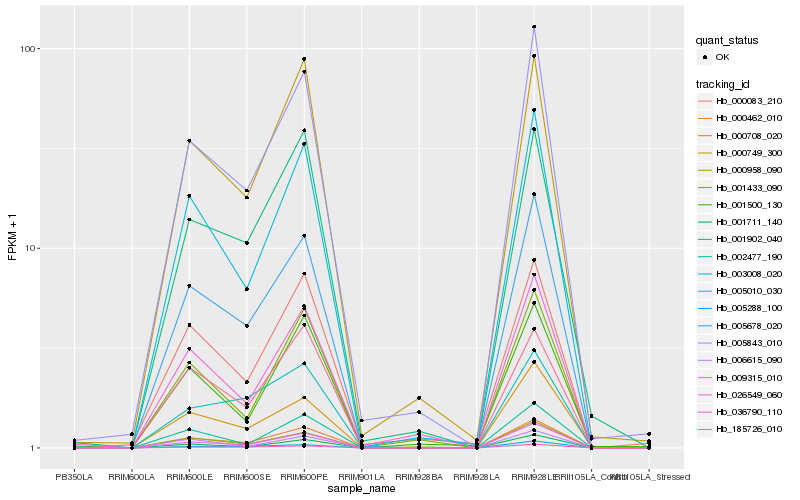
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000462_010 |
0.0 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 2 |
Hb_005678_020 |
0.0361977852 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_005010_030 |
0.0586992182 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 4 |
Hb_009315_010 |
0.0600022897 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 5 |
Hb_005843_010 |
0.0621496523 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 6 |
Hb_001711_140 |
0.0732099327 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 7 |
Hb_036790_110 |
0.081375352 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 8 |
Hb_000749_300 |
0.0818452057 |
- |
- |
PREDICTED: uncharacterized protein LOC105129180 [Populus euphratica] |
| 9 |
Hb_002477_190 |
0.0915960163 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 10 |
Hb_185726_010 |
0.0935310103 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 11 |
Hb_001902_040 |
0.09737679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000708_020 |
0.0989125098 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_000083_210 |
0.1017362227 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 14 |
Hb_000958_090 |
0.1054079335 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 15 |
Hb_001433_090 |
0.105419549 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 16 |
Hb_026549_060 |
0.1064585492 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_005288_100 |
0.1067478884 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 18 |
Hb_006615_090 |
0.1084585817 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_001500_130 |
0.110044112 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 20 |
Hb_003008_020 |
0.1102693699 |
- |
- |
auxin-responsive family protein [Populus trichocarpa] |