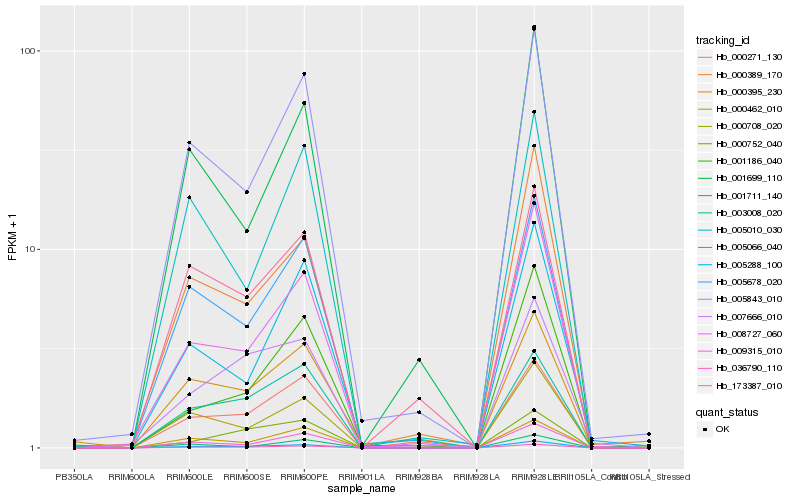
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036790_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 2 |
Hb_005288_100 |
0.0387521778 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 3 |
Hb_005678_020 |
0.0716599781 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_009315_010 |
0.0776552759 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 5 |
Hb_000462_010 |
0.081375352 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |
| 6 |
Hb_005843_010 |
0.0848273566 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_003008_020 |
0.0886465135 |
- |
- |
auxin-responsive family protein [Populus trichocarpa] |
| 8 |
Hb_008727_060 |
0.0887974234 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 9 |
Hb_000708_020 |
0.1078843881 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_000271_130 |
0.1182194346 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 11 |
Hb_000389_170 |
0.1197552317 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 12 |
Hb_000752_040 |
0.124601814 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_005010_030 |
0.1269302128 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 14 |
Hb_173387_010 |
0.1269900866 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 15 |
Hb_001711_140 |
0.1278922457 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 16 |
Hb_005066_040 |
0.1283708205 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000395_230 |
0.1297212166 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Vitis vinifera] |
| 18 |
Hb_001699_110 |
0.1307007759 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 19 |
Hb_007666_010 |
0.130720861 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 20 |
Hb_001186_040 |
0.1309125034 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |