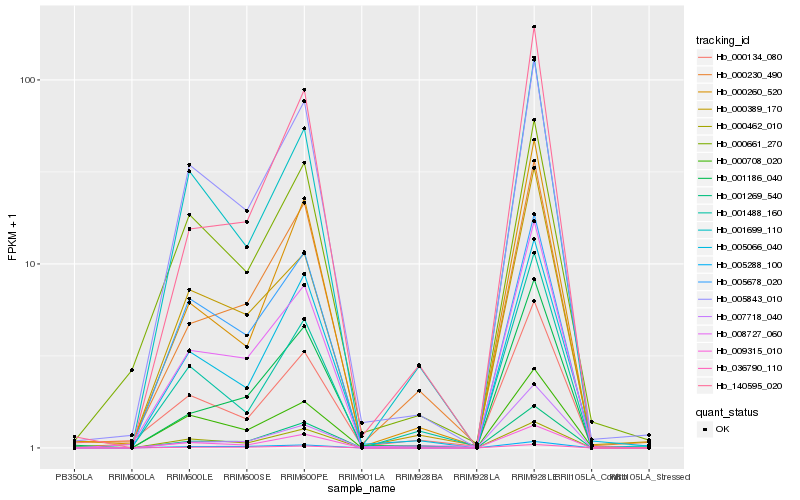
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008727_060 |
0.0 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_000389_170 |
0.0721733509 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 3 |
Hb_009315_010 |
0.0859326001 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 4 |
Hb_005288_100 |
0.0887089179 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 5 |
Hb_036790_110 |
0.0887974234 |
- |
- |
hypothetical protein JCGZ_22271 [Jatropha curcas] |
| 6 |
Hb_005843_010 |
0.0946904984 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001699_110 |
0.0987919044 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 8 |
Hb_000260_520 |
0.0994634823 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_001186_040 |
0.1025635244 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 10 |
Hb_140595_020 |
0.1038224424 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 11 |
Hb_005066_040 |
0.1099785066 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 12 |
Hb_000708_020 |
0.1123317889 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_000230_490 |
0.1151176323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005678_020 |
0.115199608 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_001488_160 |
0.1157446667 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000134_080 |
0.1164848296 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_001269_540 |
0.1166672219 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 18 |
Hb_000661_270 |
0.1254138697 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_007718_040 |
0.1255345223 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 20 |
Hb_000462_010 |
0.1268737251 |
- |
- |
hypothetical protein VITISV_041696 [Vitis vinifera] |