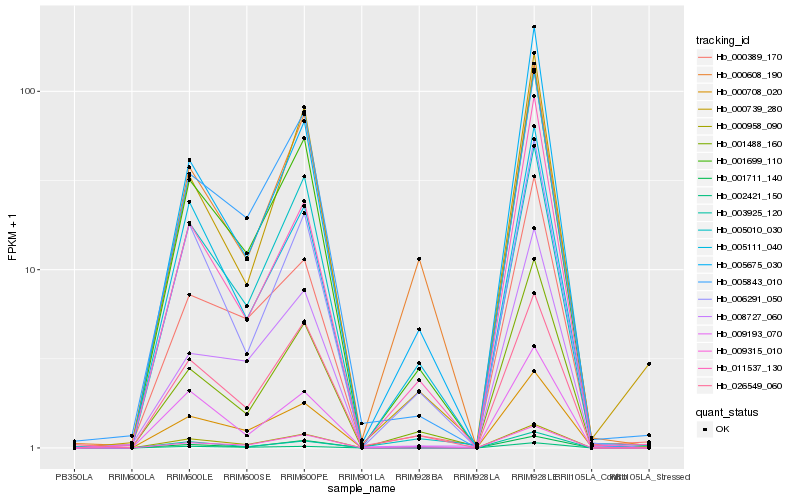
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001699_110 |
0.0 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 2 |
Hb_001488_160 |
0.0698198101 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_005675_030 |
0.077697286 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 4 |
Hb_000958_090 |
0.0793635786 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 5 |
Hb_006291_050 |
0.0821576533 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_000389_170 |
0.0847872521 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 7 |
Hb_005843_010 |
0.0935017163 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000708_020 |
0.0937318785 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_011537_130 |
0.0944420252 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_005111_040 |
0.0944704484 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 11 |
Hb_009315_010 |
0.0945309474 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 12 |
Hb_000739_280 |
0.0956336259 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_003925_120 |
0.0963939766 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_009193_070 |
0.096617345 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 15 |
Hb_008727_060 |
0.0987919044 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 16 |
Hb_001711_140 |
0.1014318788 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 17 |
Hb_026549_060 |
0.1026543322 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_000608_190 |
0.1042844831 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 19 |
Hb_002421_150 |
0.1072491808 |
- |
- |
hypothetical protein PRUPE_ppa020511mg [Prunus persica] |
| 20 |
Hb_005010_030 |
0.1073388656 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |