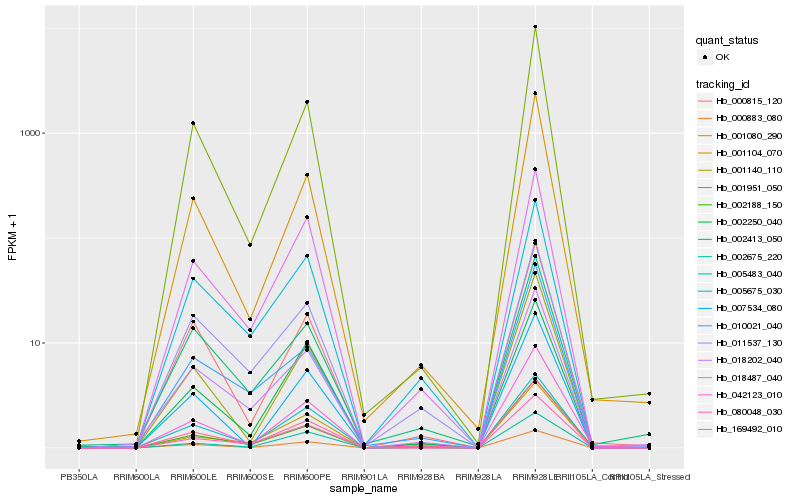
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169492_010 |
0.0 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 2 |
Hb_080048_030 |
0.0668485624 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 3 |
Hb_018202_040 |
0.0696304886 |
- |
- |
PREDICTED: phosphoenolpyruvate/phosphate translocator 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_005675_030 |
0.0717553645 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 5 |
Hb_018487_040 |
0.073365104 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 6 |
Hb_011537_130 |
0.0755936668 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001951_050 |
0.0811660833 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 8 |
Hb_001104_070 |
0.0826630957 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 9 |
Hb_000815_120 |
0.0853717566 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_002188_150 |
0.0857251906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002675_220 |
0.0860473198 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 12 |
Hb_001080_290 |
0.0884330216 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 13 |
Hb_000883_080 |
0.0922099759 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_010021_040 |
0.0929253201 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 15 |
Hb_005483_040 |
0.0942978016 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 16 |
Hb_001140_110 |
0.095216767 |
- |
- |
hypothetical protein JCGZ_26123 [Jatropha curcas] |
| 17 |
Hb_007534_080 |
0.0952450499 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_042123_010 |
0.095384807 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Populus euphratica] |
| 19 |
Hb_002250_040 |
0.0960902961 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 20 |
Hb_002413_050 |
0.0967284541 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |