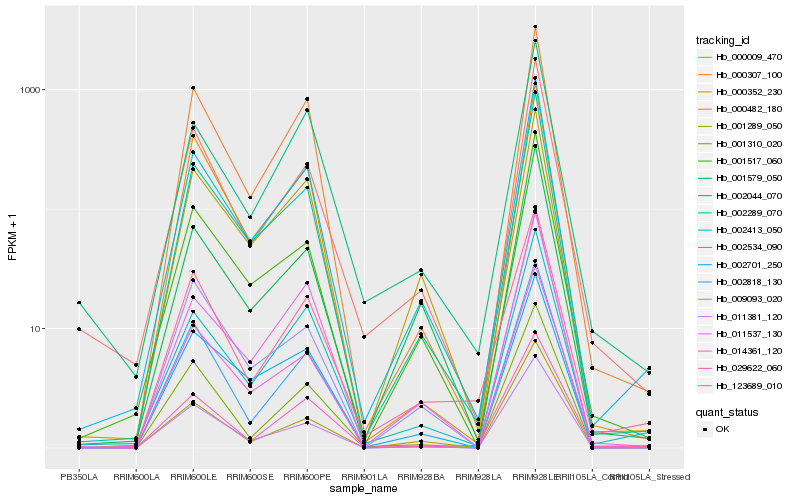
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_090 |
0.0 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 2 |
Hb_002289_070 |
0.0419444262 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002413_050 |
0.052363728 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 4 |
Hb_001517_060 |
0.0676419225 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 5 |
Hb_011537_130 |
0.0716794706 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000009_470 |
0.0771165464 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_011381_120 |
0.0776155944 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 8 |
Hb_000307_100 |
0.0792607259 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_014361_120 |
0.0805290733 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 10 |
Hb_029622_060 |
0.084700124 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_123689_010 |
0.0847381511 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 12 |
Hb_001579_050 |
0.0849701653 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_002701_250 |
0.088245033 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 14 |
Hb_009093_020 |
0.0885972986 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 15 |
Hb_001310_020 |
0.0889758586 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_002044_070 |
0.0892125284 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 17 |
Hb_001289_050 |
0.0898970349 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 18 |
Hb_000482_180 |
0.0906366157 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 19 |
Hb_002818_130 |
0.0936637802 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000352_230 |
0.0946657301 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |