| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
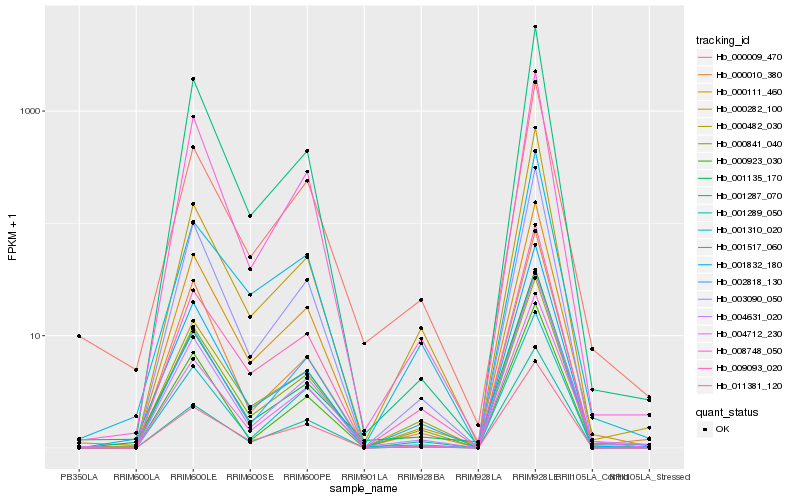
| 1 |
Hb_000841_040 |
0.0 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003090_050 |
0.0362042264 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 3 |
Hb_009093_020 |
0.0412099327 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 4 |
Hb_000111_460 |
0.0533134796 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 5 |
Hb_011381_120 |
0.0548453094 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 6 |
Hb_001832_180 |
0.0549860104 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 7 |
Hb_004631_020 |
0.0610155253 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 8 |
Hb_000282_100 |
0.0657066125 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 9 |
Hb_001287_070 |
0.0683289473 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 10 |
Hb_001289_050 |
0.0695432811 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 11 |
Hb_008748_050 |
0.0746757579 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_000010_380 |
0.0747079811 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 13 |
Hb_000923_030 |
0.0752244289 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 14 |
Hb_004712_230 |
0.0771512799 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000482_030 |
0.0777069834 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 16 |
Hb_001135_170 |
0.0788682284 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 17 |
Hb_001517_060 |
0.0792695741 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 18 |
Hb_001310_020 |
0.082407968 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_000009_470 |
0.0833543672 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_002818_130 |
0.0846323748 |
- |
- |
cytochrome P450, putative [Ricinus communis] |