| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
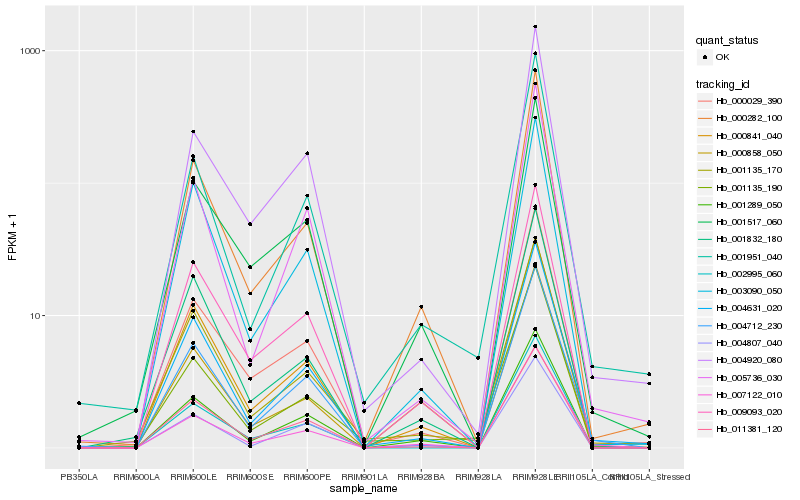
Hb_000282_100 |
0.0 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 2 |
Hb_001289_050 |
0.0585144986 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 3 |
Hb_009093_020 |
0.0625038444 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 4 |
Hb_000029_390 |
0.0650799442 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000841_040 |
0.0657066125 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_004631_020 |
0.0676305478 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_001832_180 |
0.0739619135 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 8 |
Hb_001135_170 |
0.0800923748 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 9 |
Hb_007122_010 |
0.0813253826 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 10 |
Hb_000858_050 |
0.0815087713 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_001951_040 |
0.0835331495 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_003090_050 |
0.0857588753 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 13 |
Hb_002995_060 |
0.0869167192 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 14 |
Hb_001135_190 |
0.0871670335 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 15 |
Hb_001517_060 |
0.0872387136 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 16 |
Hb_004712_230 |
0.0900611577 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 17 |
Hb_004920_080 |
0.0913792255 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_004807_040 |
0.0914110011 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 19 |
Hb_005736_030 |
0.0915876133 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 20 |
Hb_011381_120 |
0.0918155342 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |