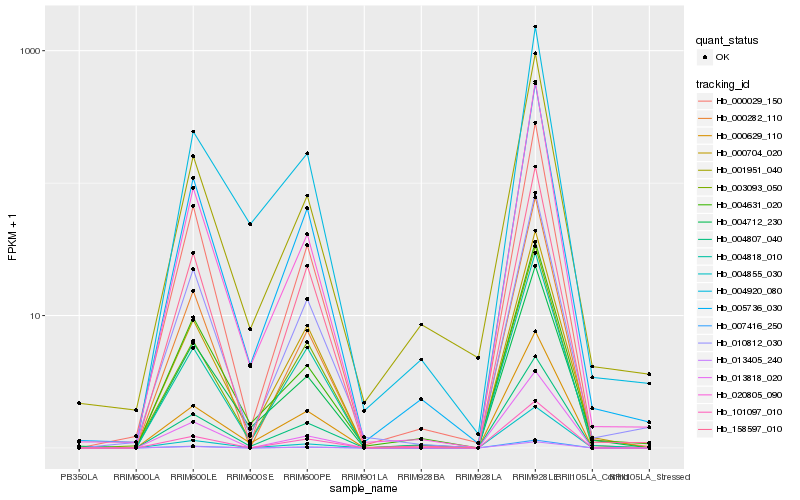
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005736_030 |
0.0 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 2 |
Hb_000029_150 |
0.0490355732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_020805_090 |
0.0635888302 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_004712_230 |
0.0635999142 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000629_110 |
0.065298246 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 6 |
Hb_004631_020 |
0.065307108 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_004807_040 |
0.0669721903 |
- |
- |
PREDICTED: uncharacterized protein LOC104420559 [Eucalyptus grandis] |
| 8 |
Hb_013405_240 |
0.0682939944 |
- |
- |
Hydroxyproline-rich glycoprotein family protein [Theobroma cacao] |
| 9 |
Hb_007416_250 |
0.0683805195 |
- |
- |
- |
| 10 |
Hb_001951_040 |
0.0686111935 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000282_110 |
0.0693716497 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |
| 12 |
Hb_101097_010 |
0.0695765045 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_004818_010 |
0.0696255344 |
- |
- |
hypothetical protein JCGZ_13628 [Jatropha curcas] |
| 14 |
Hb_000704_020 |
0.0703399808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010812_030 |
0.0704061275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_158597_010 |
0.0732739011 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_003093_050 |
0.0735446113 |
- |
- |
PREDICTED: uncharacterized protein LOC105641023 isoform X2 [Jatropha curcas] |
| 18 |
Hb_013818_020 |
0.075805793 |
- |
- |
hypothetical protein POPTR_0005s21070g [Populus trichocarpa] |
| 19 |
Hb_004855_030 |
0.0799637567 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_004920_080 |
0.0801039589 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |