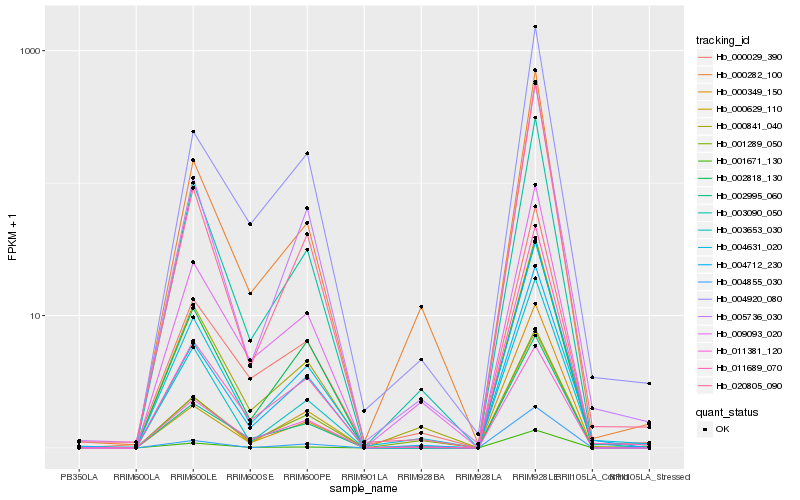
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002995_060 |
0.0 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 2 |
Hb_000029_390 |
0.0599403536 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004920_080 |
0.0620593328 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000629_110 |
0.0719361796 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 5 |
Hb_011381_120 |
0.0729815205 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 6 |
Hb_004712_230 |
0.0733662037 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001671_130 |
0.0758657339 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 8 |
Hb_009093_020 |
0.0808474608 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 9 |
Hb_020805_090 |
0.0835029478 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_004631_020 |
0.0861712636 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 11 |
Hb_000282_100 |
0.0869167192 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 12 |
Hb_005736_030 |
0.0873005011 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 13 |
Hb_000841_040 |
0.0940441856 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001289_050 |
0.0941795645 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 15 |
Hb_002818_130 |
0.0942132496 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000349_150 |
0.0963721218 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_003653_030 |
0.0968114936 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_011689_070 |
0.0984445119 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004855_030 |
0.0985999581 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_003090_050 |
0.0989155378 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |