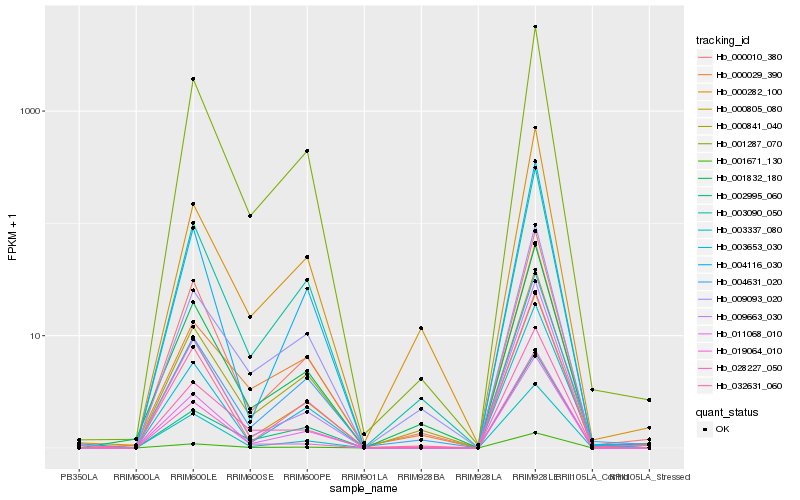
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001671_130 |
0.0 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 2 |
Hb_028227_050 |
0.0500706092 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_009663_030 |
0.0625696347 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 4 |
Hb_002995_060 |
0.0758657339 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 5 |
Hb_001287_070 |
0.0790944938 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 6 |
Hb_019064_010 |
0.0845662823 |
- |
- |
- |
| 7 |
Hb_001832_180 |
0.0848494608 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 8 |
Hb_011068_010 |
0.0864467447 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 9 |
Hb_003653_030 |
0.0910174075 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_003337_080 |
0.0932687908 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_000029_390 |
0.0938125387 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000282_100 |
0.0957511587 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 13 |
Hb_000010_380 |
0.0978526423 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 14 |
Hb_000841_040 |
0.0981503717 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_003090_050 |
0.098872857 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 16 |
Hb_004116_030 |
0.099014831 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 17 |
Hb_032631_060 |
0.0991170234 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 18 |
Hb_009093_020 |
0.0999255552 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 19 |
Hb_004631_020 |
0.1015313158 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_000805_080 |
0.101994385 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |