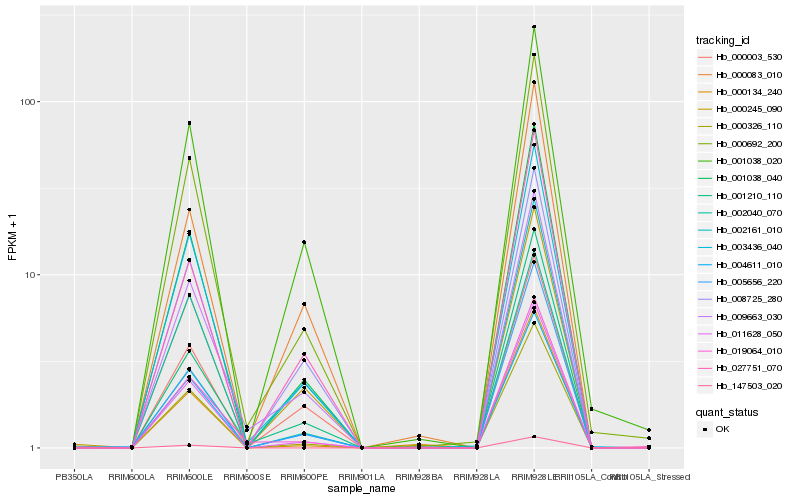
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001038_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_011628_050 |
0.0421422162 |
- |
- |
PREDICTED: monofunctional riboflavin biosynthesis protein RIBA 3, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000692_200 |
0.0438257586 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003436_040 |
0.0522851519 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 5 |
Hb_005656_220 |
0.0581311454 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 6 |
Hb_004611_010 |
0.0625745093 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 7 |
Hb_002161_010 |
0.0638310676 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 8 |
Hb_027751_070 |
0.0663541444 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000326_110 |
0.0687335361 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 10 |
Hb_002040_070 |
0.0698681788 |
- |
- |
- |
| 11 |
Hb_008725_280 |
0.07238951 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 12 |
Hb_000083_010 |
0.0733908918 |
- |
- |
- |
| 13 |
Hb_001210_110 |
0.0751570032 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 14 |
Hb_000245_090 |
0.075376221 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 15 |
Hb_009663_030 |
0.0760636939 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 16 |
Hb_000003_530 |
0.078324907 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 17 |
Hb_019064_010 |
0.0788107151 |
- |
- |
- |
| 18 |
Hb_001038_020 |
0.0797438458 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 19 |
Hb_000134_240 |
0.0798849401 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_147503_020 |
0.0901622307 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |