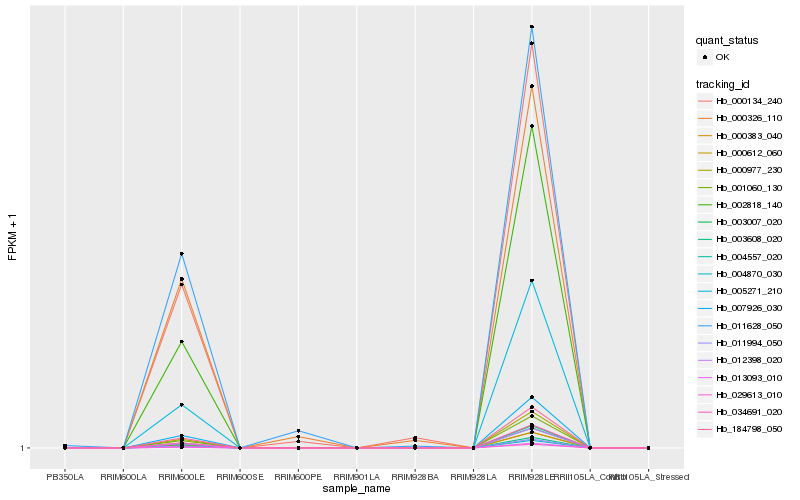
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_240 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000326_110 |
0.0541005957 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 3 |
Hb_011628_050 |
0.0650521819 |
- |
- |
PREDICTED: monofunctional riboflavin biosynthesis protein RIBA 3, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_004557_020 |
0.0773515059 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 5 |
Hb_012398_020 |
0.0773533042 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 6 |
Hb_003007_020 |
0.0773580125 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 7 |
Hb_013093_010 |
0.0773654091 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 8 |
Hb_034691_020 |
0.0773681749 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 9 |
Hb_029613_010 |
0.0773791164 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_003608_020 |
0.0773832172 |
- |
- |
hypothetical protein CISIN_1g041945mg [Citrus sinensis] |
| 11 |
Hb_000383_040 |
0.0774136177 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 12 |
Hb_000612_060 |
0.0774185665 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 13 |
Hb_004870_030 |
0.0775876069 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_011994_050 |
0.0776932716 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 15 |
Hb_001060_130 |
0.0777405693 |
- |
- |
- |
| 16 |
Hb_000977_230 |
0.078147728 |
- |
- |
FT-like protein [Betula luminifera] |
| 17 |
Hb_005271_210 |
0.0784489578 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 18 |
Hb_007926_030 |
0.0785884149 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP13-like [Populus euphratica] |
| 19 |
Hb_184798_050 |
0.0787220314 |
- |
- |
- |
| 20 |
Hb_002818_140 |
0.0790558511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |