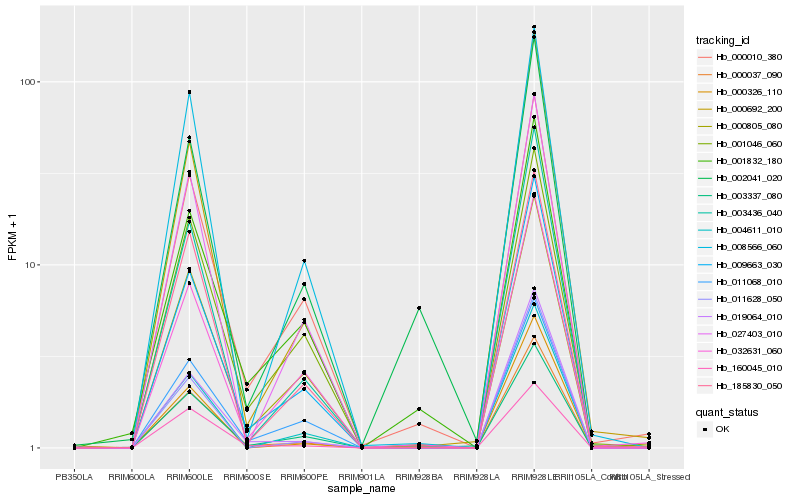
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000037_090 |
0.0 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 2 |
Hb_019064_010 |
0.0674318468 |
- |
- |
- |
| 3 |
Hb_009663_030 |
0.0704859563 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 4 |
Hb_003436_040 |
0.0748696062 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 5 |
Hb_000326_110 |
0.0758204514 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 6 |
Hb_003337_080 |
0.0831775734 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 7 |
Hb_000010_380 |
0.0844444236 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 8 |
Hb_002041_020 |
0.0885075958 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 9 |
Hb_000692_200 |
0.0888630637 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 10 |
Hb_185830_050 |
0.0890432944 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 11 |
Hb_001832_180 |
0.0901199873 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 12 |
Hb_004611_010 |
0.0905735768 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 13 |
Hb_027403_010 |
0.0927643409 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 14 |
Hb_000805_080 |
0.0948448367 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011628_050 |
0.0956214871 |
- |
- |
PREDICTED: monofunctional riboflavin biosynthesis protein RIBA 3, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_160045_010 |
0.0974999598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008566_060 |
0.0979018563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011068_010 |
0.1006731711 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 19 |
Hb_001046_060 |
0.1007840929 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_032631_060 |
0.1026693883 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |