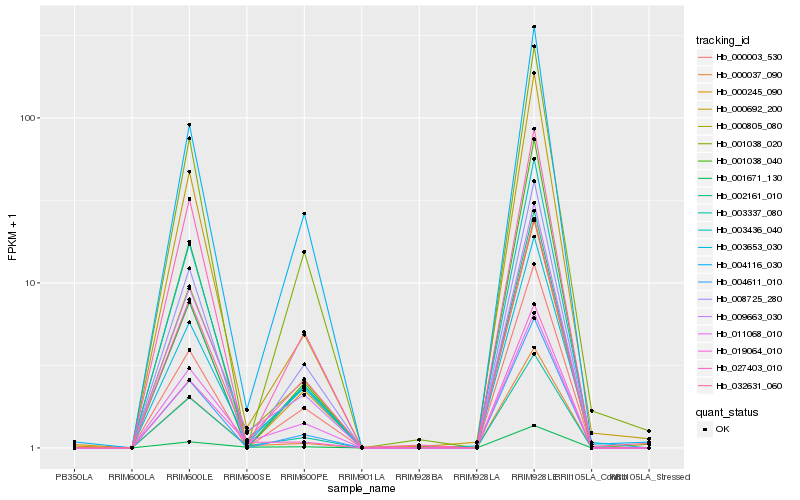
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009663_030 |
0.0 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 2 |
Hb_003436_040 |
0.0550127769 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 3 |
Hb_003337_080 |
0.0595454632 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 4 |
Hb_004611_010 |
0.060150879 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 5 |
Hb_001671_130 |
0.0625696347 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 6 |
Hb_000692_200 |
0.063728239 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 7 |
Hb_008725_280 |
0.0652606524 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 8 |
Hb_002161_010 |
0.0665360754 |
- |
- |
PREDICTED: 3,9-dihydroxypterocarpan 6A-monooxygenase-like [Jatropha curcas] |
| 9 |
Hb_019064_010 |
0.0667836383 |
- |
- |
- |
| 10 |
Hb_000245_090 |
0.0693383685 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 11 |
Hb_004116_030 |
0.0695667934 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 12 |
Hb_032631_060 |
0.0704479565 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 13 |
Hb_000037_090 |
0.0704859563 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 14 |
Hb_003653_030 |
0.0706443897 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_011068_010 |
0.0707135963 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 16 |
Hb_000805_080 |
0.0744204575 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_027403_010 |
0.0748334496 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 18 |
Hb_001038_020 |
0.075161844 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 19 |
Hb_001038_040 |
0.0760636939 |
- |
- |
- |
| 20 |
Hb_000003_530 |
0.0761294036 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |