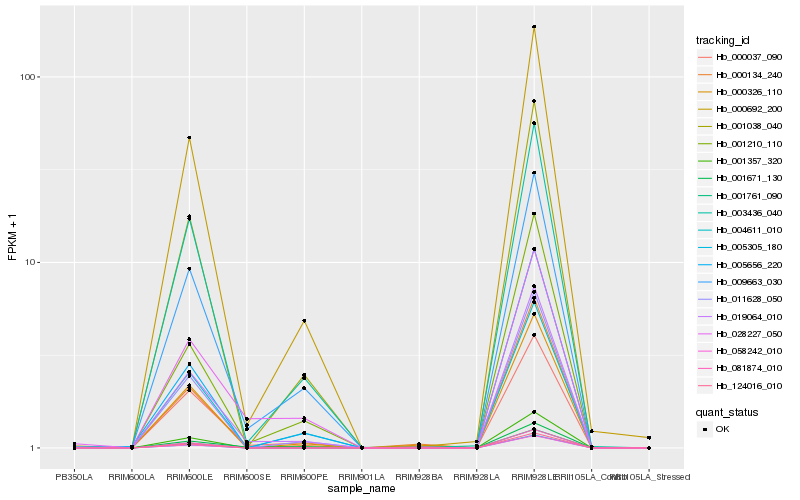
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019064_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_009663_030 |
0.0667836383 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 3 |
Hb_000037_090 |
0.0674318468 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 4 |
Hb_000692_200 |
0.0677319207 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 5 |
Hb_011628_050 |
0.0759015504 |
- |
- |
PREDICTED: monofunctional riboflavin biosynthesis protein RIBA 3, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_003436_040 |
0.0768226712 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 7 |
Hb_001038_040 |
0.0788107151 |
- |
- |
- |
| 8 |
Hb_000326_110 |
0.0820706496 |
- |
- |
PREDICTED: uncharacterized protein LOC105632350 [Jatropha curcas] |
| 9 |
Hb_001671_130 |
0.0845662823 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 10 |
Hb_001210_110 |
0.0908332482 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 11 |
Hb_000134_240 |
0.0913887616 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_028227_050 |
0.1024235293 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_004611_010 |
0.1026235201 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 14 |
Hb_005656_220 |
0.1028357265 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 15 |
Hb_001357_320 |
0.1042475687 |
- |
- |
GRP3S, putative [Ricinus communis] |
| 16 |
Hb_124016_010 |
0.1042667839 |
- |
- |
hypothetical protein POPTR_0008s21806g [Populus trichocarpa] |
| 17 |
Hb_081874_010 |
0.1043763043 |
- |
- |
PREDICTED: uncharacterized protein LOC102581051 [Solanum tuberosum] |
| 18 |
Hb_001761_090 |
0.1043865769 |
- |
- |
Protein STRUBBELIG-RECEPTOR FAMILY 5 [Glycine soja] |
| 19 |
Hb_005305_180 |
0.1045498586 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010-like [Gossypium raimondii] |
| 20 |
Hb_058242_010 |
0.1046317762 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |