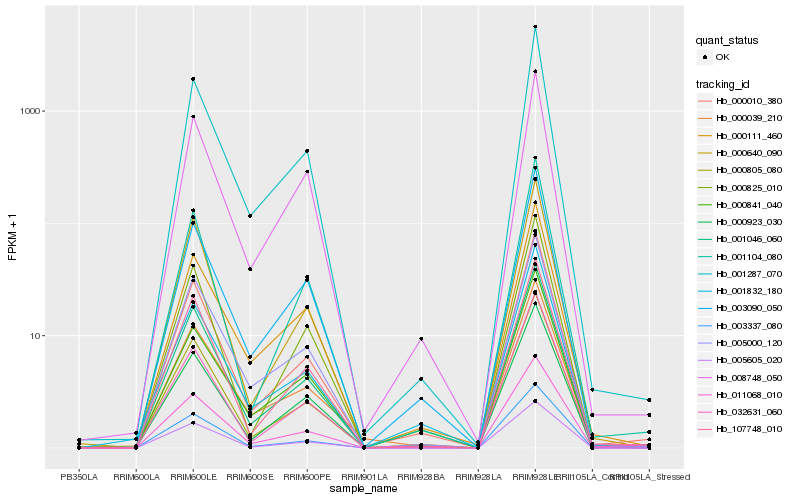
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_380 |
0.0 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 2 |
Hb_000805_080 |
0.0342551743 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001046_060 |
0.0410394958 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 4 |
Hb_001287_070 |
0.0462580027 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 5 |
Hb_032631_060 |
0.0488152334 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 6 |
Hb_011068_010 |
0.0532193979 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 7 |
Hb_003337_080 |
0.0541452791 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 8 |
Hb_003090_050 |
0.0551139784 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 9 |
Hb_001104_080 |
0.056940524 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 10 |
Hb_000923_030 |
0.0574945231 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 11 |
Hb_005605_020 |
0.0636996957 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 12 |
Hb_001832_180 |
0.0650404337 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 13 |
Hb_005000_120 |
0.0655662908 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 14 |
Hb_107748_010 |
0.0665989382 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000640_090 |
0.0713673494 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_008748_050 |
0.0727444864 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_000825_010 |
0.073081346 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 18 |
Hb_000039_210 |
0.0733435404 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 19 |
Hb_000841_040 |
0.0747079811 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_000111_460 |
0.0761353315 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |