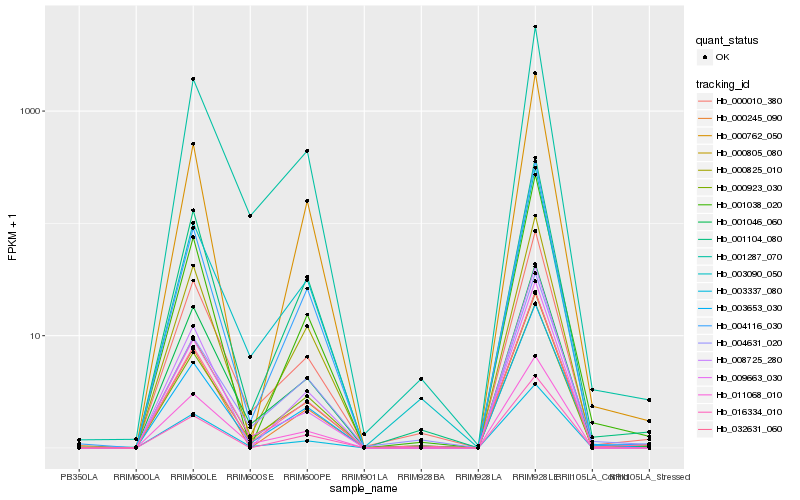
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032631_060 |
0.0 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 2 |
Hb_001104_080 |
0.0396429324 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 3 |
Hb_000010_380 |
0.0488152334 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 4 |
Hb_004116_030 |
0.0515963139 |
- |
- |
Chlorophyll a-b binding CP29.3, chloroplastic -like protein [Gossypium arboreum] |
| 5 |
Hb_000805_080 |
0.0524251532 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000923_030 |
0.0526106395 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 7 |
Hb_003653_030 |
0.0553014894 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001038_020 |
0.0574264124 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 9 |
Hb_000825_010 |
0.0602057757 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 10 |
Hb_001287_070 |
0.0645942451 |
- |
- |
latex plastidic aldolase-like protein [Hevea brasiliensis] |
| 11 |
Hb_004631_020 |
0.0666917321 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 12 |
Hb_008725_280 |
0.06712543 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s19920g [Populus trichocarpa] |
| 13 |
Hb_003090_050 |
0.06814504 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 14 |
Hb_000762_050 |
0.0692725502 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000245_090 |
0.0694942632 |
- |
- |
PREDICTED: uncharacterized protein LOC105631554 [Jatropha curcas] |
| 16 |
Hb_009663_030 |
0.0704479565 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 17 |
Hb_003337_080 |
0.070515191 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 18 |
Hb_011068_010 |
0.070861295 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 19 |
Hb_016334_010 |
0.0709887497 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 20 |
Hb_001046_060 |
0.0710594391 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |