| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
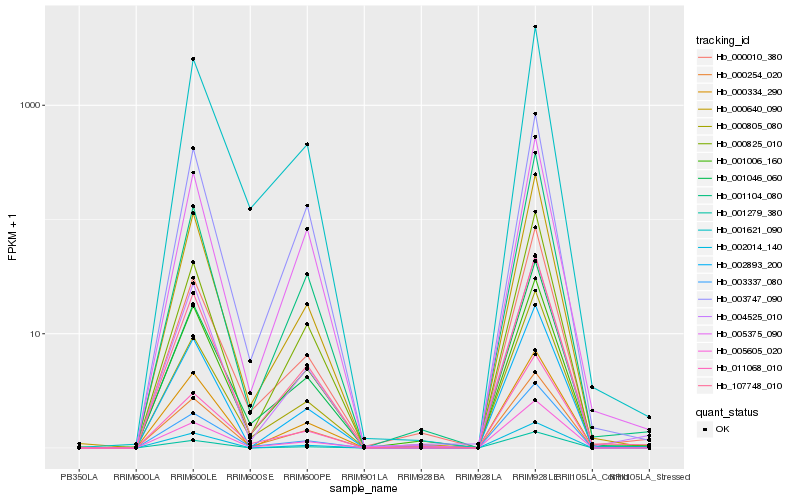
Hb_107748_010 |
0.0 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000640_090 |
0.0356422328 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_005605_020 |
0.0407799772 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 4 |
Hb_001046_060 |
0.0408321175 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 5 |
Hb_002893_200 |
0.0448655307 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000805_080 |
0.0534339703 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001104_080 |
0.060157427 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 8 |
Hb_000825_010 |
0.0604457921 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 9 |
Hb_001621_090 |
0.0611588941 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 10 |
Hb_004525_010 |
0.0615966274 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 11 |
Hb_005375_090 |
0.0621602186 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001006_160 |
0.0635661057 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_003337_080 |
0.0637064282 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 14 |
Hb_003747_090 |
0.0645951866 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_002014_140 |
0.0649183472 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 16 |
Hb_011068_010 |
0.0650837851 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 17 |
Hb_000254_020 |
0.0663818166 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 18 |
Hb_000010_380 |
0.0665989382 |
- |
- |
hypothetical protein CICLE_v10020897mg [Citrus clementina] |
| 19 |
Hb_001279_380 |
0.0671002308 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 20 |
Hb_000334_290 |
0.0673262821 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |