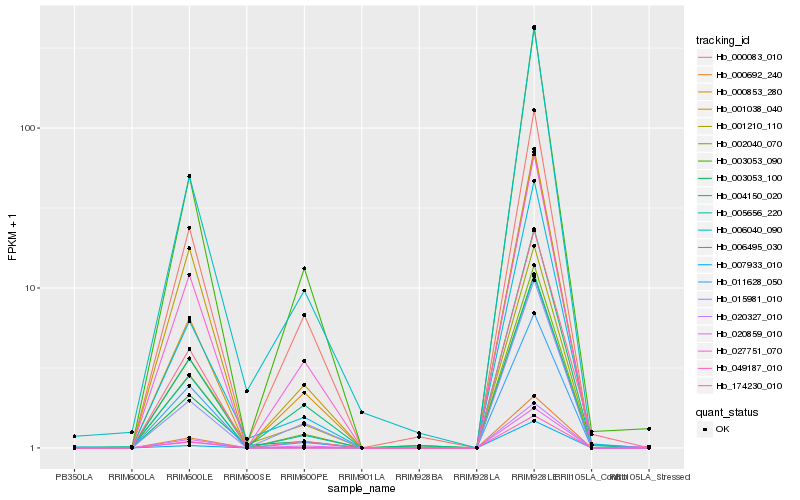
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002040_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_005656_220 |
0.0407861296 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 3 |
Hb_001210_110 |
0.0441182412 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1 [Jatropha curcas] |
| 4 |
Hb_003053_090 |
0.051545131 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_027751_070 |
0.051954116 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_020859_010 |
0.0547727126 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X6 [Populus euphratica] |
| 7 |
Hb_006495_030 |
0.0561900551 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 8 |
Hb_006040_090 |
0.0618028205 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Hevea brasiliensis] |
| 9 |
Hb_001038_040 |
0.0698681788 |
- |
- |
- |
| 10 |
Hb_004150_020 |
0.0720642291 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 11 |
Hb_000083_010 |
0.0731656861 |
- |
- |
- |
| 12 |
Hb_000853_280 |
0.076237005 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 13 |
Hb_003053_100 |
0.0768862114 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 14 |
Hb_015981_010 |
0.0806227736 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_174230_010 |
0.08130402 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 16 |
Hb_007933_010 |
0.0827076231 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_020327_010 |
0.0854234144 |
- |
- |
PREDICTED: uncharacterized protein LOC104886355, partial [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000692_240 |
0.0855109985 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_049187_010 |
0.0855461091 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 20 |
Hb_011628_050 |
0.0884464398 |
- |
- |
PREDICTED: monofunctional riboflavin biosynthesis protein RIBA 3, chloroplastic isoform X1 [Jatropha curcas] |