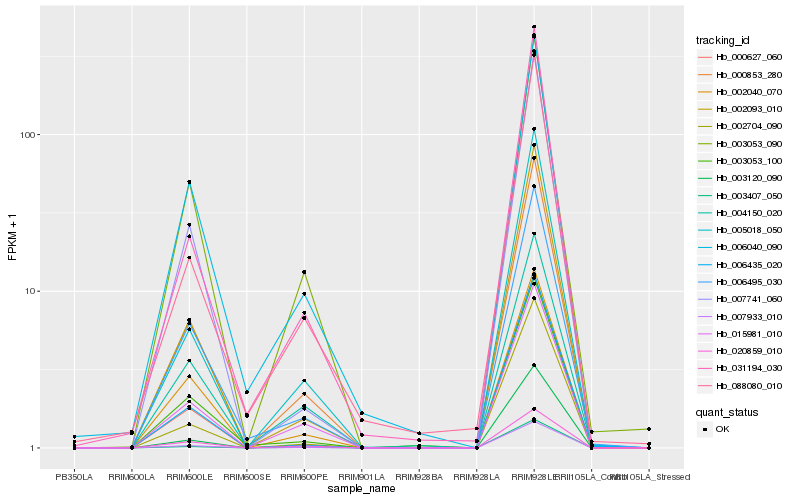
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_280 |
0.0 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 2 |
Hb_007933_010 |
0.0114171469 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 3 |
Hb_003120_090 |
0.0409004968 |
- |
- |
PREDICTED: uncharacterized protein LOC100264374 [Vitis vinifera] |
| 4 |
Hb_002093_010 |
0.0459081758 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 5 |
Hb_002704_090 |
0.0552668362 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_005018_050 |
0.0577833905 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 7 |
Hb_000627_060 |
0.058456301 |
- |
- |
- |
| 8 |
Hb_003053_090 |
0.0606981851 |
- |
- |
PREDICTED: carbonic anhydrase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_015981_010 |
0.061147925 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_003407_050 |
0.0643633327 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_006495_030 |
0.0652468067 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 12 |
Hb_007741_060 |
0.0656627269 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 13 |
Hb_006435_020 |
0.0663389667 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_031194_030 |
0.0676945316 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 15 |
Hb_006040_090 |
0.0688523314 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Hevea brasiliensis] |
| 16 |
Hb_020859_010 |
0.0702201581 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X6 [Populus euphratica] |
| 17 |
Hb_003053_100 |
0.0707725606 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 18 |
Hb_088080_010 |
0.0742120303 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 19 |
Hb_002040_070 |
0.076237005 |
- |
- |
- |
| 20 |
Hb_004150_020 |
0.0785646228 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |