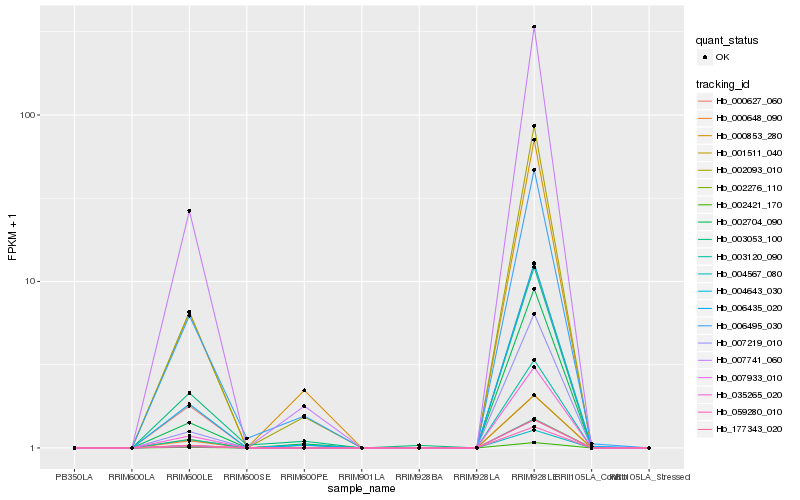
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007741_060 |
0.0 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 2 |
Hb_000627_060 |
0.0160334731 |
- |
- |
- |
| 3 |
Hb_002093_010 |
0.0294702061 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 4 |
Hb_006435_020 |
0.0328183953 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_177343_020 |
0.0328268153 |
- |
- |
PREDICTED: uncharacterized protein LOC104751876 [Camelina sativa] |
| 6 |
Hb_004643_030 |
0.0330481759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004567_080 |
0.0330564416 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_035265_020 |
0.0369462008 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000648_090 |
0.043043801 |
- |
- |
- |
| 10 |
Hb_002704_090 |
0.0476443405 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_002276_110 |
0.0487955921 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_002421_170 |
0.0492260548 |
- |
- |
hypothetical protein POPTR_0003s18240g [Populus trichocarpa] |
| 13 |
Hb_059280_010 |
0.0509602367 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 14 |
Hb_007219_010 |
0.056554331 |
- |
- |
- |
| 15 |
Hb_007933_010 |
0.0568402424 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 16 |
Hb_001511_040 |
0.0596526688 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 17 |
Hb_000853_280 |
0.0656627269 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 18 |
Hb_003120_090 |
0.0712970819 |
- |
- |
PREDICTED: uncharacterized protein LOC100264374 [Vitis vinifera] |
| 19 |
Hb_003053_100 |
0.0741403935 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 20 |
Hb_006495_030 |
0.0809428971 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |