| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
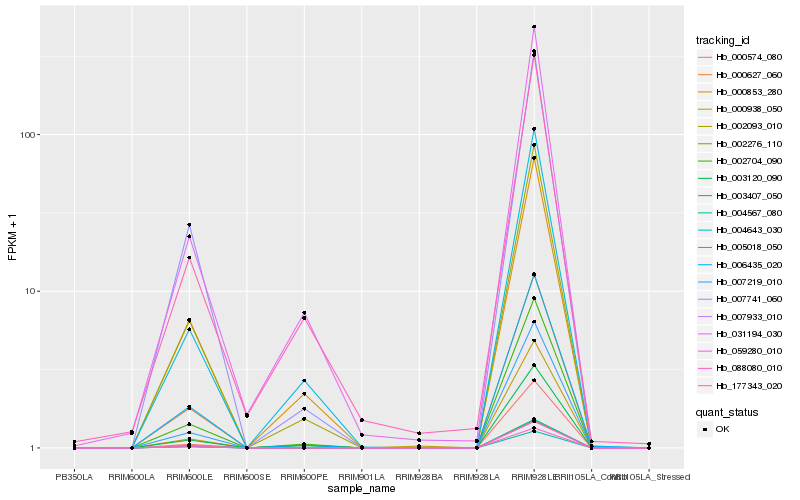
Hb_002704_090 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_002093_010 |
0.022569932 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 3 |
Hb_000627_060 |
0.0318165297 |
- |
- |
- |
| 4 |
Hb_003120_090 |
0.0325153935 |
- |
- |
PREDICTED: uncharacterized protein LOC100264374 [Vitis vinifera] |
| 5 |
Hb_005018_050 |
0.0394662172 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 6 |
Hb_007933_010 |
0.0443319216 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_031194_030 |
0.0460195422 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 8 |
Hb_007741_060 |
0.0476443405 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 9 |
Hb_006435_020 |
0.0476876734 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 10 |
Hb_003407_050 |
0.0513686974 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000853_280 |
0.0552668362 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 12 |
Hb_002276_110 |
0.0580904406 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_059280_010 |
0.0581170797 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 14 |
Hb_007219_010 |
0.0587876693 |
- |
- |
- |
| 15 |
Hb_004567_080 |
0.0666195682 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 16 |
Hb_004643_030 |
0.0666500285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000938_050 |
0.0682089529 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |
| 18 |
Hb_088080_010 |
0.0698210845 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 19 |
Hb_177343_020 |
0.0700070721 |
- |
- |
PREDICTED: uncharacterized protein LOC104751876 [Camelina sativa] |
| 20 |
Hb_000574_080 |
0.0708031127 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |