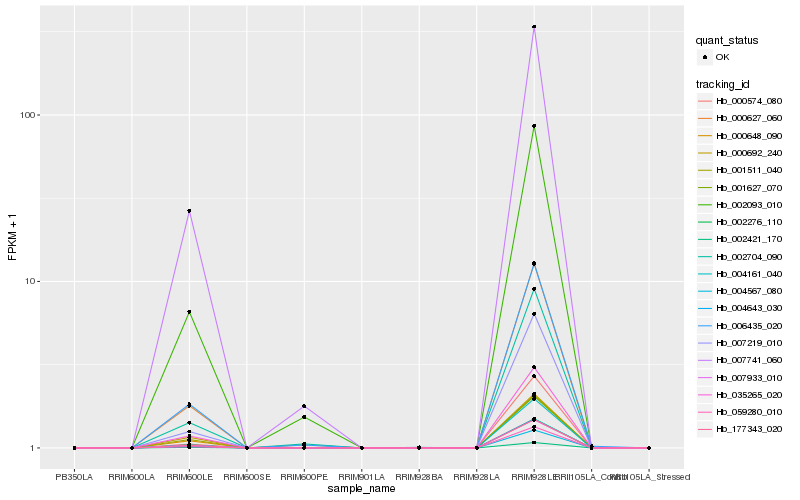
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004567_080 |
0.0 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_004643_030 |
6.24446e-05 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_177343_020 |
0.006488032 |
- |
- |
PREDICTED: uncharacterized protein LOC104751876 [Camelina sativa] |
| 4 |
Hb_035265_020 |
0.0214970943 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002276_110 |
0.0317829135 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000648_090 |
0.0323372588 |
- |
- |
- |
| 7 |
Hb_007741_060 |
0.0330564416 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 8 |
Hb_059280_010 |
0.034658399 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 9 |
Hb_002421_170 |
0.0411589747 |
- |
- |
hypothetical protein POPTR_0003s18240g [Populus trichocarpa] |
| 10 |
Hb_007219_010 |
0.041733662 |
- |
- |
- |
| 11 |
Hb_000627_060 |
0.0421957785 |
- |
- |
- |
| 12 |
Hb_006435_020 |
0.0505964996 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 13 |
Hb_001511_040 |
0.0542667104 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 14 |
Hb_002093_010 |
0.0551408413 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 15 |
Hb_002704_090 |
0.0666195682 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000574_080 |
0.0729069266 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 17 |
Hb_004161_040 |
0.0771886527 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Populus euphratica] |
| 18 |
Hb_000692_240 |
0.0811577972 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_007933_010 |
0.0815182627 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 20 |
Hb_001627_070 |
0.0816297884 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1 [Nelumbo nucifera] |