| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
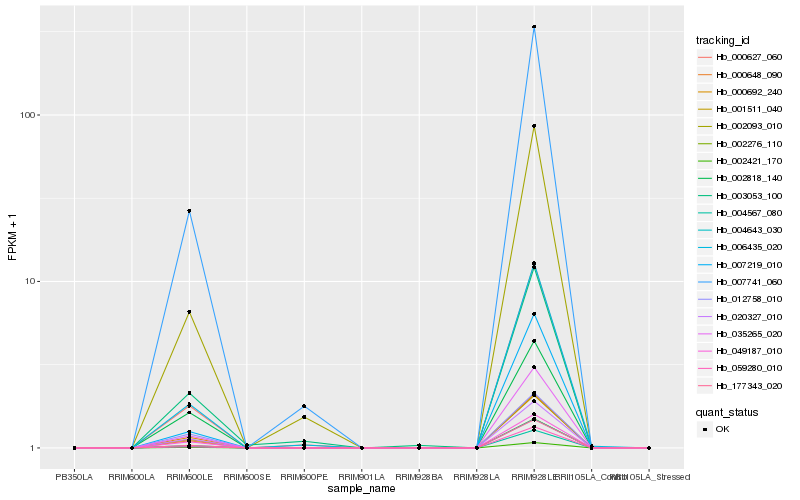
Hb_002421_170 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s18240g [Populus trichocarpa] |
| 2 |
Hb_000648_090 |
0.0088444975 |
- |
- |
- |
| 3 |
Hb_001511_040 |
0.0131612388 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 4 |
Hb_035265_020 |
0.0196966611 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_177343_020 |
0.0346902433 |
- |
- |
PREDICTED: uncharacterized protein LOC104751876 [Camelina sativa] |
| 6 |
Hb_000692_240 |
0.0402260048 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_004643_030 |
0.0410967543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004567_080 |
0.0411589747 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 9 |
Hb_020327_010 |
0.0439931757 |
- |
- |
PREDICTED: uncharacterized protein LOC104886355, partial [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_049187_010 |
0.0486865657 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 11 |
Hb_007741_060 |
0.0492260548 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 12 |
Hb_000627_060 |
0.0640857661 |
- |
- |
- |
| 13 |
Hb_006435_020 |
0.0671102908 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_002276_110 |
0.0727202092 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002093_010 |
0.0747783758 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 16 |
Hb_059280_010 |
0.0755640307 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 17 |
Hb_012758_010 |
0.0809160974 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 2 [Jatropha curcas] |
| 18 |
Hb_002818_140 |
0.0809868906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007219_010 |
0.0825522583 |
- |
- |
- |
| 20 |
Hb_003053_100 |
0.0835411674 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Jatropha curcas] |