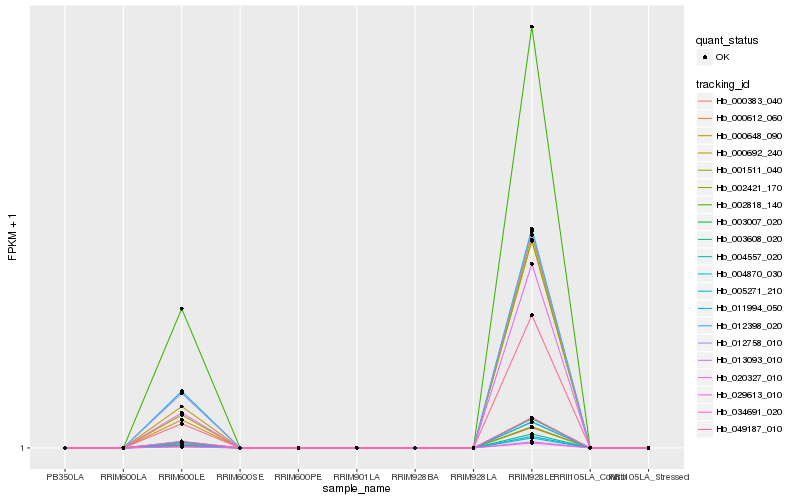
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020327_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104886355, partial [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000692_240 |
0.00377631 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_049187_010 |
0.0047068999 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 4 |
Hb_001511_040 |
0.0308580344 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 5 |
Hb_012758_010 |
0.0370868717 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 2 [Jatropha curcas] |
| 6 |
Hb_002818_140 |
0.0371580963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005271_210 |
0.0404128542 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 8 |
Hb_002421_170 |
0.0439931757 |
- |
- |
hypothetical protein POPTR_0003s18240g [Populus trichocarpa] |
| 9 |
Hb_011994_050 |
0.0462107569 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_004870_030 |
0.0474403587 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_000612_060 |
0.0502557553 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 12 |
Hb_000383_040 |
0.0503759335 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 13 |
Hb_029613_010 |
0.0513956574 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_013093_010 |
0.0519790587 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 15 |
Hb_000648_090 |
0.0528059794 |
- |
- |
- |
| 16 |
Hb_012398_020 |
0.0528289261 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 17 |
Hb_004557_020 |
0.0530813386 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 18 |
Hb_003007_020 |
0.0545963087 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 19 |
Hb_034691_020 |
0.0551679604 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 20 |
Hb_003608_020 |
0.055762308 |
- |
- |
hypothetical protein CISIN_1g041945mg [Citrus sinensis] |