| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
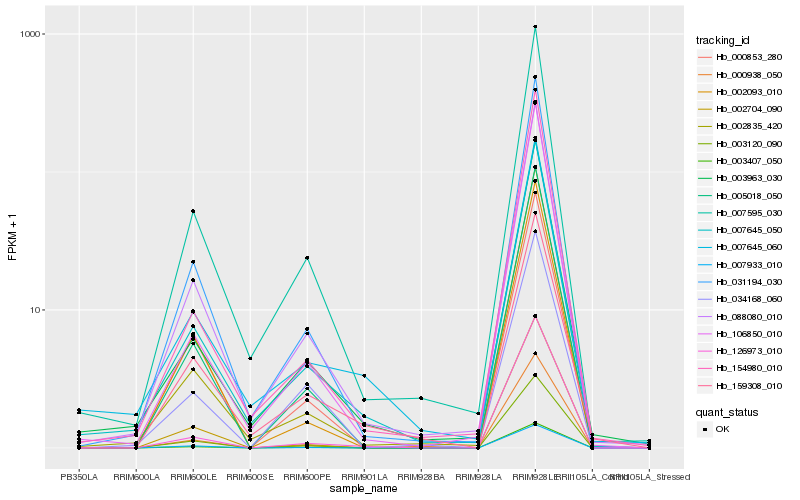
Hb_007595_030 |
0.0 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 2 |
Hb_088080_010 |
0.0205330697 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 3 |
Hb_007645_050 |
0.0383932755 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 4 |
Hb_031194_030 |
0.0386367982 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 5 |
Hb_003963_030 |
0.0507253564 |
- |
- |
psbB [Jatropha curcas] |
| 6 |
Hb_005018_050 |
0.0581646631 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 7 |
Hb_003407_050 |
0.0592718341 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_003120_090 |
0.0616807566 |
- |
- |
PREDICTED: uncharacterized protein LOC100264374 [Vitis vinifera] |
| 9 |
Hb_002835_420 |
0.0668534226 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 10 |
Hb_154980_010 |
0.0676035831 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 11 |
Hb_000938_050 |
0.0740044237 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |
| 12 |
Hb_002704_090 |
0.0762041119 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_106850_010 |
0.0763377952 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 14 |
Hb_007933_010 |
0.0775217284 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_000853_280 |
0.0798303049 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 16 |
Hb_126973_010 |
0.0804332594 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 17 |
Hb_007645_060 |
0.0841511813 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 18 |
Hb_159308_010 |
0.0843362773 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 19 |
Hb_002093_010 |
0.0854791681 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 20 |
Hb_034168_060 |
0.0874872255 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |