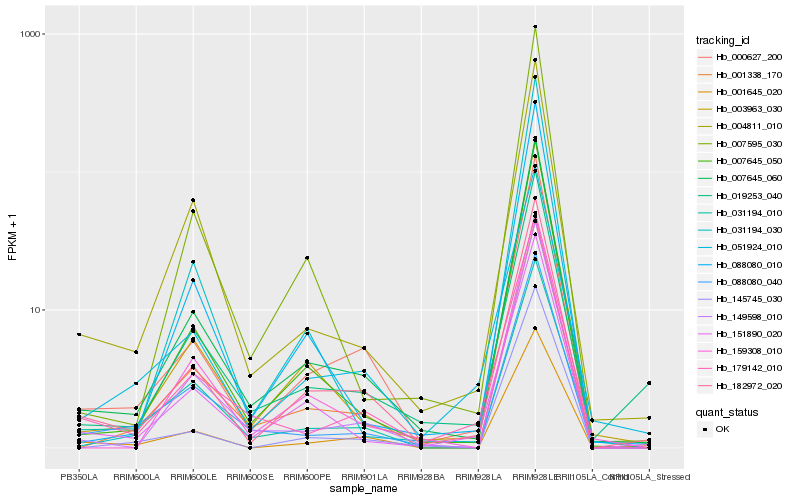
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007645_060 |
0.0 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 2 |
Hb_007645_050 |
0.0613647508 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 3 |
Hb_182972_020 |
0.0680202784 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 4 |
Hb_149598_010 |
0.06914476 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 5 |
Hb_003963_030 |
0.0709836622 |
- |
- |
psbB [Jatropha curcas] |
| 6 |
Hb_000627_200 |
0.0721789276 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 7 |
Hb_001338_170 |
0.075524053 |
- |
- |
NADH dehydrogenase subunit 4 [Hevea brasiliensis] |
| 8 |
Hb_151890_020 |
0.0793870626 |
- |
- |
photosystem II cp47 protein, partial (chloroplast) [Pentaphragma sp. Duangjai 49] |
| 9 |
Hb_088080_010 |
0.0828075208 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 10 |
Hb_004811_010 |
0.0837830559 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 11 |
Hb_007595_030 |
0.0841511813 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 12 |
Hb_159308_010 |
0.0887804769 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 13 |
Hb_088080_040 |
0.0896923335 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 2 A [Medicago truncatula] |
| 14 |
Hb_179142_010 |
0.0962291498 |
- |
- |
NADH dehydrogenase ND 2 subunit [Nicotiana tomentosiformis] |
| 15 |
Hb_019253_040 |
0.0962669231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_145745_030 |
0.0974478151 |
- |
- |
BnaC09g26890D [Brassica napus] |
| 17 |
Hb_031194_030 |
0.1031840655 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 18 |
Hb_031194_010 |
0.104692523 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 19 |
Hb_001645_020 |
0.1047084793 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit H [Medicago truncatula] |
| 20 |
Hb_051924_010 |
0.1065264168 |
- |
- |
BnaA01g34100D [Brassica napus] |