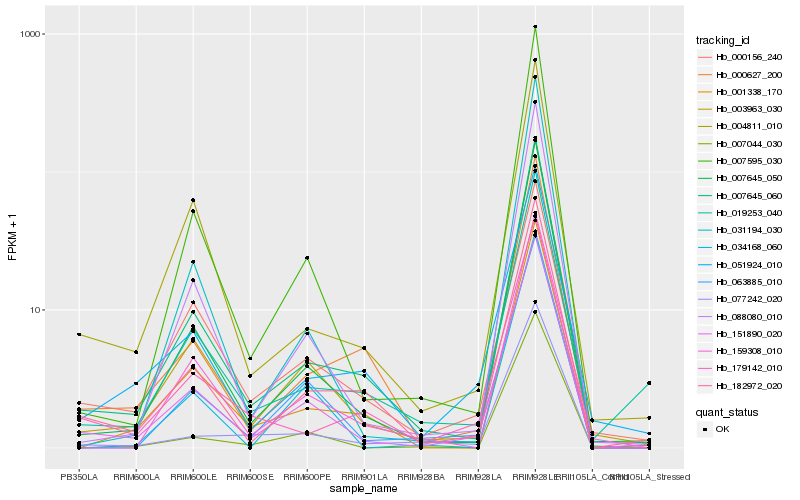
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_151890_020 |
0.0 |
- |
- |
photosystem II cp47 protein, partial (chloroplast) [Pentaphragma sp. Duangjai 49] |
| 2 |
Hb_007645_060 |
0.0793870626 |
- |
- |
cytochrome f [Hevea brasiliensis] |
| 3 |
Hb_003963_030 |
0.0846647796 |
- |
- |
psbB [Jatropha curcas] |
| 4 |
Hb_007645_050 |
0.0857821248 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 5 |
Hb_182972_020 |
0.0870673602 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 6 |
Hb_007595_030 |
0.0905018777 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 7 |
Hb_088080_010 |
0.0926967088 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 8 |
Hb_063885_010 |
0.1027929556 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_004811_010 |
0.1029708714 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 10 |
Hb_019253_040 |
0.1132394489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007044_030 |
0.1132621488 |
- |
- |
Os06g0273400 [Oryza sativa Japonica Group] |
| 12 |
Hb_000627_200 |
0.1137604636 |
- |
- |
NADH dehydrogenase subunit I [Hevea brasiliensis] |
| 13 |
Hb_031194_030 |
0.1149731572 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 14 |
Hb_001338_170 |
0.1157103071 |
- |
- |
NADH dehydrogenase subunit 4 [Hevea brasiliensis] |
| 15 |
Hb_077242_020 |
0.1159184026 |
- |
- |
ribosomal protein S4 (mitochondrion) [Ricinus communis] |
| 16 |
Hb_051924_010 |
0.1162210637 |
- |
- |
BnaA01g34100D [Brassica napus] |
| 17 |
Hb_034168_060 |
0.1169320982 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |
| 18 |
Hb_159308_010 |
0.1183719462 |
- |
- |
ribosomal protein S4 [Cercidiphyllum japonicum] |
| 19 |
Hb_000156_240 |
0.1204241727 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 20 |
Hb_179142_010 |
0.1212439145 |
- |
- |
NADH dehydrogenase ND 2 subunit [Nicotiana tomentosiformis] |