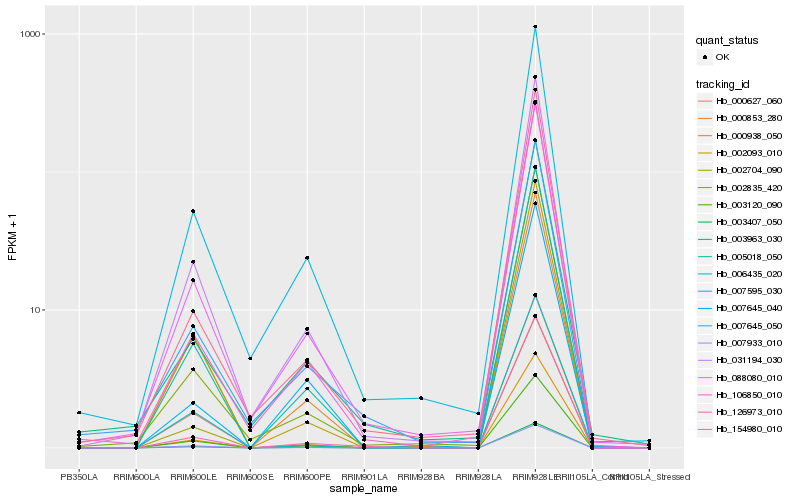
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005018_050 |
0.0 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 2 |
Hb_003407_050 |
0.019896328 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_003120_090 |
0.0206497184 |
- |
- |
PREDICTED: uncharacterized protein LOC100264374 [Vitis vinifera] |
| 4 |
Hb_031194_030 |
0.0371941414 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 5 |
Hb_002704_090 |
0.0394662172 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_007933_010 |
0.0517864627 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_088080_010 |
0.0541551967 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 8 |
Hb_002093_010 |
0.055259859 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 9 |
Hb_000853_280 |
0.0577833905 |
- |
- |
PREDICTED: extensin-like [Jatropha curcas] |
| 10 |
Hb_007595_030 |
0.0581646631 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 11 |
Hb_126973_010 |
0.0601438133 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 12 |
Hb_000938_050 |
0.06239392 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |
| 13 |
Hb_002835_420 |
0.0675124461 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 14 |
Hb_000627_060 |
0.0677189663 |
- |
- |
- |
| 15 |
Hb_154980_010 |
0.0715636303 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 16 |
Hb_007645_050 |
0.0727410368 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 17 |
Hb_106850_010 |
0.0729157781 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 18 |
Hb_006435_020 |
0.0742796506 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_003963_030 |
0.0813283266 |
- |
- |
psbB [Jatropha curcas] |
| 20 |
Hb_007645_040 |
0.0819340077 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |