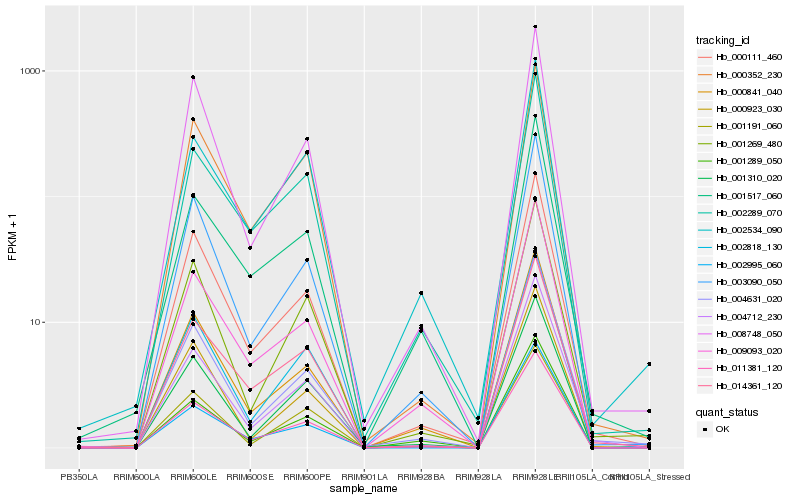
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011381_120 |
0.0 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 2 |
Hb_009093_020 |
0.0489952673 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 3 |
Hb_002818_130 |
0.0521912251 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000841_040 |
0.0548453094 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000111_460 |
0.0563584709 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 6 |
Hb_001310_020 |
0.0575217052 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_003090_050 |
0.0578371178 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 8 |
Hb_002289_070 |
0.0582803852 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004712_230 |
0.0672318067 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 10 |
Hb_014361_120 |
0.0686160034 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 11 |
Hb_002995_060 |
0.0729815205 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 12 |
Hb_004631_020 |
0.0744813336 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 13 |
Hb_001289_050 |
0.0751148119 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 14 |
Hb_001517_060 |
0.0766283854 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 15 |
Hb_000352_230 |
0.0769079226 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_001191_060 |
0.0769124306 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 17 |
Hb_002534_090 |
0.0776155944 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 18 |
Hb_008748_050 |
0.0799338635 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_001269_480 |
0.0800360867 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000923_030 |
0.0805784827 |
- |
- |
F-box family protein [Populus trichocarpa] |