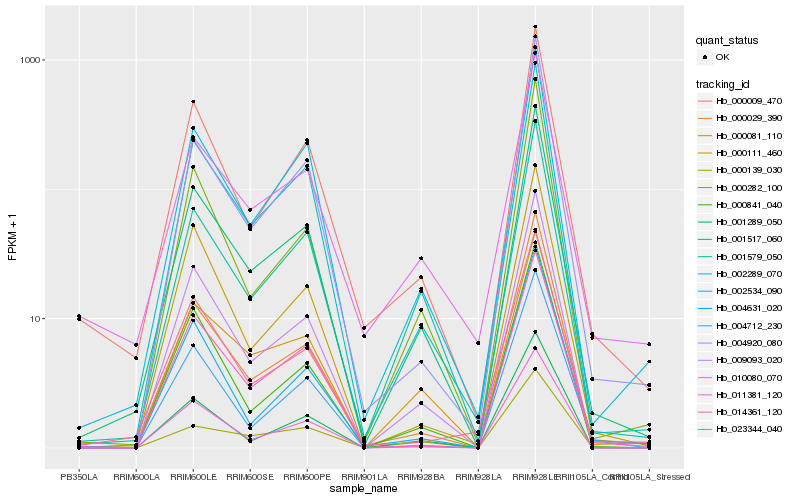
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001517_060 |
0.0 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 2 |
Hb_009093_020 |
0.0555077801 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 3 |
Hb_002289_070 |
0.0573698323 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002534_090 |
0.0676419225 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 5 |
Hb_001579_050 |
0.0708652258 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000029_390 |
0.0725670203 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000009_470 |
0.0732745675 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_011381_120 |
0.0766283854 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2-like [Jatropha curcas] |
| 9 |
Hb_000081_110 |
0.0781081288 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 10 |
Hb_001289_050 |
0.0784324496 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 11 |
Hb_000841_040 |
0.0792695741 |
transcription factor |
TF Family: C2C2-CO-like |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000282_100 |
0.0872387136 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 13 |
Hb_014361_120 |
0.0872714587 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 14 |
Hb_010080_070 |
0.0881483221 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004920_080 |
0.0902520907 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_023344_040 |
0.0910914573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000111_460 |
0.0943522481 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 18 |
Hb_004712_230 |
0.095246193 |
- |
- |
PREDICTED: retrotransposon-like protein 1 [Jatropha curcas] |
| 19 |
Hb_004631_020 |
0.0960536592 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_000139_030 |
0.0964868146 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |