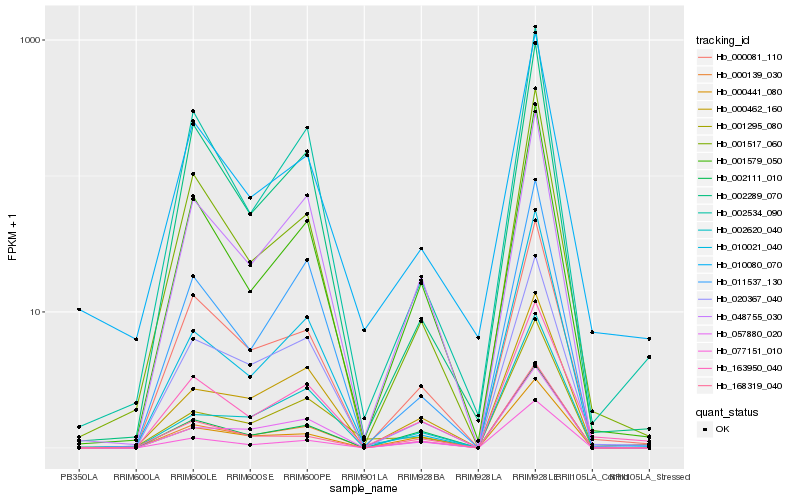
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 2 |
Hb_001579_050 |
0.0787247899 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000081_110 |
0.0832477316 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_057880_020 |
0.0838801679 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 5 |
Hb_000462_160 |
0.091002533 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_020367_040 |
0.0914744916 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |
| 7 |
Hb_048755_030 |
0.0915000899 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 8 |
Hb_000441_080 |
0.0917977935 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_002620_040 |
0.0925249609 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 10 |
Hb_001517_060 |
0.0964868146 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 11 |
Hb_002111_010 |
0.0983379612 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 12 |
Hb_163950_040 |
0.1011197272 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001295_080 |
0.1055531966 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 14 |
Hb_168319_040 |
0.1058385007 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 15 |
Hb_077151_010 |
0.1074311976 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 16 |
Hb_011537_130 |
0.1121486503 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002289_070 |
0.1123576077 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010021_040 |
0.1137588706 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 19 |
Hb_002534_090 |
0.1138455858 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 20 |
Hb_010080_070 |
0.1146288463 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |